Abstract
1184
Objectives: Many medical image processing applications rely on targeted regions of interest within a larger volumetric image. In this work, we considered the application of classifying trans-axial CT image planes into 6 main anatomical body regions: head, neck, chest, abdomen, pelvis and legs, which can then, for example, be used to describe the location of tumors in automatically generated reports. We describe and compare three competing methods with regards to implementation complexity, performance and limitations.
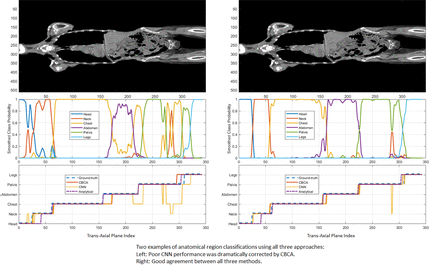
Methods: We implemented three CT plane classification methods as follows: (1) An analytical approach which doesn’t require any training and relies solely on Hounsfield unit profiles to identify cut-planes between anatomical regions, (2) an approach using convolutional neural networks (CNNs) of varying configuration (322, 642 and 1282 input image matrix sizes) to classify each plane independently, requiring training using a labeled image set, and (3) CNN followed by a context based correction algorithm (CBCA) which improves the CNN classification using positional relationships between all CT planes. 140 whole-body CT datasets were semi-automatically labeled into anatomical regions using a custom viewer that was initialized with analytical approach cut-planes and were then manually adjusted. Of these, 90 CTs (32,562 planes) were used to train the CNNs. To evaluate performance, we used the remaining 50 CT image sets (18,278 planes) and compared automated plane classification to manual labels using bootstrapping (1000 samples, 30 image each) to estimate performance variability.
Results: Mean plane labeling accuracy ± standard-error of all 50 CTs with the analytical method was 85.91±0.00%. The three CNN variant accuracies were 95.11±0.09%, 95.85±0.05% and 96.05±0.02% and in all three cases were improved by CBCA to 95.50±0.06%, 96.20±0.07% and 96.32±0.01% (p<0.001 for all), eliminating out-of-order (e.g. pelvis proximal to neck) regions errors. Allowing for ±1 cm ambiguity in anatomical region boundary definition, plane classification accuracies further improved to 90.25±0.00% for analytical; 98.80±0.08%, 99.20±0.05%, 99.22±0.03% for CNNs; and 99.10±0.04%, 99.36±0.08%, 99.43±0.01% for CBCA (p<0.001). CBCA improvement was more dramatic with unoptimized CNNs. Execution times with all methods were <10 seconds per CT image set on a standard laptop.
Conclusions: The analytical approach achieved acceptable accuracy for anatomical region segmentation without the need for explicit data labelling and was effective for batch labelling CT data sets, greatly reducing manual labeling efforts. Convolutional neural networks achieved superior accuracy and allowed for rapid development and training but required labelled data. Post-hoc correction of CNN results using an analytical context-based correction further improved classification, achieving nearly perfect CT plane labeling and anatomical region segmentation.