Abstract
Noninvasive reporter gene imaging is a component of molecular imaging. Reporter imaging can provide noninvasive assessments of endogenous biologic processes in living subjects and can be performed using different imaging modalities. This review will focus on radionuclide-based reporter gene imaging as developed and applied in preclinical and clinical studies. Examples of different reporter systems are presented, with a focus on human reporter systems. Selected applications are discussed, including adoptive cell therapies, gene and oncoviral therapies, oncogenesis, signal pathway monitoring, and imaging drug treatment. Molecular imaging, and noninvasive reporter gene imaging in particular, are making important contributions to our understanding of disease development, progression, and treatment in our current era of molecular medicine and individualized patient care.
The convergence of imaging technology and the disciplines of molecular and cell biology in the mid-1990s provided the foundation for the development of noninvasive molecular imaging using reporter genes. This development was originally stimulated by programs funded by the National Institutes of Health and the Department of Energy. This initiative has now spread globally throughout Europe and Asia, and new molecular imaging societies and new journals have been developed to address this rapidly expanding discipline.
Reporter gene imaging is only a single component of molecular imaging. Researchers developed reporter genes in the 1980s, using the bacterial β-galactosidase (1) and chloramphenicol acetyltransferase (2) genes, to study various cellular processes both in vitro and in vivo. However, visualization of these reporter enzymes required postmortem tissue sampling and processing (1,2). Advances in cell biology eventually stimulated the development of novel noninvasive visualization systems that could provide accurate and sensitive measurements in animal models of human disease.
Noninvasive molecular imaging in living animals is the direct result of significant developments in several imaging technologies, including MR imaging (3), radionuclide imaging (quantitative autoradiography, γ-camera imaging, and PET) (4), and optical imaging of small animals (5). These developments occurred more or less in parallel to each other and were largely independent of the advances that were occurring in genetics and in molecular and cell biology during the 1980s and early 1990s. Each of these imaging technologies also had important antecedents. For example, the development of radionuclide-based molecular imaging is founded on the radiotracer principle, which was first described by George de Hevesy. In 1935 he published a letter in Nature on the tracer principle using 32P (6), and he was awarded the Nobel Prize in chemistry in 1943. The tracer technique was later adapted for many applications in physiology and biochemistry, as well as in functional diagnosis and in nuclear medicine and molecular imaging.
REPORTER GENE IMAGING PARADIGMS
A common feature of all reporter constructs is that the complementary DNA expression cassette, containing the reporter transgenes of interest (e.g., herpes simplex virus 1 thymidine kinase [HSV1-tk]), can be placed under the control of specific promoter–enhancer control elements. The versatility of reporter constructs is due in part to their modular design, since arrangements in the expression cassette can be varied to some extent. For example, reporter genes can be constructed to be “always on” using constitutive promoters (such as LTR, RSV, CMV, PGK, and EF1). Alternatively, the promoter–enhancer elements can be constructed to be inducible and sensitive to activation and regulation by specific endogenous transcription factors and promoters (factors that bind to and activate specific enhancer elements in the regulatory region of the reporter construct, leading to the initiation of reporter gene transcription). These strategies have been widely applied in both optical imaging (7,8) and radionuclide-based imaging (9) and to a lesser degree in MR imaging (3).
MODALITIES
In Vivo Optical Imaging
Optical reporter proteins can emit light in the visible range (300–600 nm), either through interaction with specific substrates (bioluminescence) or after excitation with light of a specific wavelength (fluorescence). The emitted light is captured by a sensitive charge-coupled-device camera (10). Both bioluminescence and fluorescence reporter genes have advantages and disadvantages. Bioluminescence imaging is the most affordable and cost-effective real-time analysis of gene expression in small animals; bioluminescence imaging has excellent sensitivity and is user-friendly. Until recently, tomographic images and volumetric semiquantification was limited because of light scatter, differential light absorption, and lack of depth information. Fluorescence reporter genes were used extensively in the past to image molecular events in single live cells but are less adaptable to imaging live animals—although issues related to autofluorescence are being addressed using spectral unmixing techniques (11). Bioluminescence imaging is more sensitive for small-animal imaging than is fluorescence imaging, because of high signal-to-background ratios and absence of autoluminescence (12). Although optical imaging provides outstanding sensitivity or resolution, it is limited in translation to clinical applications (13).
MR Imaging
Compared with radionuclide or optical reporter genes, the development and application of MR reporters is more limited (3). Three classes of MR reporter genes, based on their interaction at the molecular level, have been described (14): enzyme-based cleavage of functional groups that block water (proton) exchange, protein binding, or MR imaging contrast agents; the expression of surface receptors that enable the binding of specific MR imaging contrast agents; and the expression of para- and antiferromagnetic proteins involved with iron metabolism, such as tyrosinase and ferritin. MR imaging is probably the least used reporter gene imaging technique for small animals; it has comparatively low sensitivity but higher resolution than radionuclide imaging. MR-based reporter gene imaging has not occupied center stage, and no clinical studies have been reported.
Radionuclide Imaging
The radionuclide imaging class of reporter genes encodes for proteins that interact with and trap radiolabeled molecules, resulting in localized accumulation of radioactivity that reflects the expression level of the reporter gene. One of the first noninvasive reporter gene imaging paradigms was based on HSV1-tk and was described in 1995 (15). The HSV1-tk reporter system is a radiotracer enzymatic assay; it is similar to the FDG–hexokinase paradigm that is widely used to image glucose utilization. Other reporter strategies use a transporter or a receptor to selectively trap a radiolabeled probe, such that the level of accumulated radioactivity reflects the expression of the reporter gene. All these reporter systems involve the coupling of a reporter gene with a specific reporter substrate—a complementary pair. Radionuclide-based reporters have the potential for immediate clinical application and are the focus of current development.
HSV1-tk and Other Enzyme-Based Genes
The initial and most widely used radionuclide-based reporter system is HSV1-tk (15) coupled to 124I/18F-2′-fluoro-2′-deoxy-1-β-d-arabinofuranosyl-5-iodouracil (FIAU) or 18F-9-[4-fluoro-3-(hydroxymethyl)butyl]guanine (FHBG). Unlike mammalian TK, the HSV1-tk and HSV1-tksr39 enzymes are much less substrate-specific; they phosphorylate modified thymidine analogs (e.g., FIAU and FHBG), as well as antiviral drugs (acyclovir, ganciclovir, and penciclovir). HSV1-tk had been used as a suicide therapeutic gene well before its use as a reporter gene. The Varicella zoster virus thymidine kinase gene and bicyclic nucleoside probes represent a relatively new and not fully validated PET reporter system (16).
Human Reporter Gene Systems
Recently, enzyme-based human reporter genes have been developed and validated to avoid potential immunologic reactions to HSV1-tk protein. They include human mitochondrial thymidine kinase-2 (hmtk2) (17) and several mutants (18), as well as a mutant of human deoxycytidine kinase (hdCK) using clinically available pyrimidine-based radiotracers (19). Other reporter systems have also been described: NIS (rat sodium iodide symporter (20)), hNET (human norepinephrine transporter (21)), hSSTR2 (human somatostatin receptors (22)), hCEA (human carcinoembryonic antigen [CEA] (23)), and D2R (rat dopamine D2 receptor (24)).
hmtk2/hΔTK2
A human Δ-mitochondrial thymidine kinase type 2 (hΔTK2) was developed and can be imaged with pyrimidine-based radiotracers. However, cells expressing hΔTK2 show relatively low levels of pyrimidine radiotracer accumulation compared with HSV1-tk (one sixth to one tenth) (17). Further enzyme engineering resulted in the development of a mutant hTK2-N93D/L109F with the ability to phosphorylate 18F-FMAU (18).
hdCK
hdCK was developed as a PET reporter gene. hdCK is capable of converting deoxycytidine, deoxyadenosine, deoxyguanosine, and several pyrimidine-based cytotoxic drugs (including cytosine-arabinoside and 2′-difluorocytosine) into their monophosphate forms. hdCK along with 18F-2′-deoxy-2′-fluoroarabinofuranosylcytosine was developed as a new reporter system, and hdCK mutants were shown to phosphorylate pyrimidine-based radiotracers at levels comparable to that of wild-type HSV1-tk (19). The in vitro radiotracer accumulation and in vivo imaging studies demonstrated the advantage of this mutant reporter gene over its native predecessor, because of a thymidine-permissive conformation of this enzyme that allows phosphorylation of FIAU and FEAU.
hNIS
hNIS is an intrinsic transmembrane glycoprotein and member of the sodium/solute symporter family (25). hNIS is naturally expressed in the thyroid gland and stomach and, to a lesser extent, in salivary and lactating mammary glands, the choroid plexus, kidney epithelial cells, and the placenta (20). The hNIS has many advantages as a reporter gene, primarily because of the wide availability of substrates already approved for clinical use (both for γ-camera [123I−, 131I−, and 99mTcO4−] and PET [124I− and 94mTcO4−] imaging studies), a well-understood metabolism and clearance of its substrates from the body, and the fact that radiochemistry laboratory facilities do not need to be nearby. In addition, all nuclear medicine departments have access to a γ-camera or a SPECT system (and many have PET systems). Nevertheless, hNIS also has limitations; high background activity in the thyroid, stomach, intestine, and other tissues can obscure reporter imaging (limit the sensitivity and specificity) in or adjacent to these tissues, and iodide and pertechnetate rapidly clear from hNIS-expressing cells and tissues as medium and blood levels fall.
hNET
hNET is a transmembrane protein, is one of several monoamine transporters involved in the transport of norepinephrine, epinephrine, and dopamine (26), and functions as a rapid reuptake system located at or near presynaptic terminals. Several radiolabeled probes are used clinically (123I, 131I, 123/124I-metaiodobenzylguanidine, and 11C-ephedrine), and the small size of the hNET reporter gene cassette allows it to be easily incorporated into the delivery vehicle (27). The successful imaging of hNET expression in neuroendocrine tumors and altered sympathetic enervation of the heart have led to the suggested application of hNET as a human reporter gene (21,28). Sequential imaging of animals bearing both hNET reporter-transduced and wild-type xenografts clearly showed the advantage of late imaging (27). The background-corrected radioactivity profile of the hNET-transduced xenograft shows a peak value at 8 h and a slow exponential washout (half-life, 63 h), compared with the more rapid washout from the nontransduced xenograft (half-life, 12 h). Calculating the fraction of measured radioactivity that represents trapped (nonbackground) radioactivity shows that measured values can be misleading, particularly at early times (at 1, 2, and 8 h, the trapped radioactivity is ∼14%, 52%, and 83% of the measured radioactivity, respectively). The radioactivity ratio, comparing transduced-to-nontransduced xenografts, is also highly time-dependent (increasing from 1.2:1 at 1 h to 6.5:1, 14:1, and 28:1 at 8, 24 and 48 h, respectively, after the injection of 124I-metaiodobenzylguanidine). These results highlight the importance of the imaging time when single-time experiments are compared (27).
hSSTR2
The somatostatin receptors are G-protein–linked transmembrane pass receptors. The expression of 1 of the 6 SSTr genes, subtype 2, is largely restricted to the pituitary, although other tissues express low levels of this protein (29). Although the hSSTr2 gene was originally described as a promising reporter gene in 1999 (29) and again in 2005 (30), image quality remained suboptimal using 111In-diethylenetriaminepentaacetic acid–octreotide, 99mTc-P829, 188Re-P829, 99mTc-P2045, or 94mTc-demotate-1 radioligands. This situation has now changed significantly with the availability of 68Ga-DOTATOC, 68Ga-DOTATATE, and 111In-DOTABASS, due to high specific uptake and comparatively rapid body clearance and low background activity. These peptide–chelate conjugates were originally designed for radiotherapy of SSTr2-expressing tumors. DOTATOC and DOTATATE have been classified as receptor agonists, whereas DOTABASS is an antagonist. They have undergone extensive clinical testing in both Europe (31) and the United States (32) and are being developed as clinical imaging and targeted radiotherapeutic agents.
Because ligand binding to the hSSTr2 expressed on transduced cells could potentially perturb the biology of these cells through G-protein–coupled receptor signaling (as was shown for the D2R (24,33)), this issue must be addressed. In vitro, activation of SSTr2 on tumor cells results in cell growth arrest, whereas activation of SSTr5 results in cell proliferation (34). We observed that SSTr2 transduction had variable effects on xenograft growth rates; these effects varied with cell type but not with the level of SSTr2 expression (35).
hCEA
A recombinant reporter gene based on the CEA was developed for imaging. The CEA-based reporter gene can be detected by molecular imaging using radiolabeled anti-CEA antibodies as a reporter probe. In vivo small-animal PET using a 124I-labeled anti-CEA scFv-Fc antibody visualized CEA-transfected Jurkat T cell xenografts (23).
D2R
D2R is also a potential reporter gene because of the availability of 18F-fluoroethylspiperone, a well-studied probe for PET imaging of the dopaminergic system in humans. However, only the rat D2R reporter system has been studied, and potential problems such as occupancy of the receptor by endogenous natural ligands and adverse biologic effects on transduced cells have been raised. To address this concern, the group at the University of California at Los Angeles developed a signaling-inert mutant variant (D2R80A) as a reporter gene and visualized gene expression with 18F-fluoroethylspiperone (24). They showed that the D2R80A variant has in vitro 3H-spiperone binding parameters similar to those of wild-type D2R, but there was complete uncoupling of ligand binding from activation of G-protein–linked signaling. Unfortunately, the nonhuman origin of the dopamine D2R80A receptors does not favor their use in clinical reporter gene imaging studies, and human hD2R equivalents will have to be developed.
Comparisons
Every reporter gene has its advantages and disadvantages. HSV1-tk has a greater potential to elicit an immune reaction than human reporter systems. The expression of a transporter or a receptor on the surface of all cells involves a complex process of protein trafficking and membrane integration that may affect reporter readout and quantitation. An enzyme-based reporter would be expected to produce a higher signal than a receptor-based reporter because of enzymatic or catalytic trapping of the radiolabeled substrate, resulting in signal amplification. On the other hand, intracellular enzyme–based reporters may be limited by transport of the radiolabeled substrate into transduced cells. For transporter and receptor-based reporter systems, the ligand-binding domain is located on the external plasma membrane (27) and does not require radioligand permeation of the cell membrane to bind to the receptor.
SELECTED APPLICATIONS FOR CLINICAL TRANSLATION
Molecular imaging paradigms involving reporter genes, although first applied to noninvasive in vivo imaging of small animals, are now being translated into clinical imaging paradigms. Although only 5 human reporter gene studies have been reported (36–40), translation is expected to expand and establish new standards of medical practice in the future. However, reporter gene imaging studies will always be more limited in patients than in animals, because of the need to transduce the target tissue or cells with specific reporter constructs or to produce transgenic animals bearing the reporter constructs.
Gene and Oncoviral Therapy: Clinical Trials
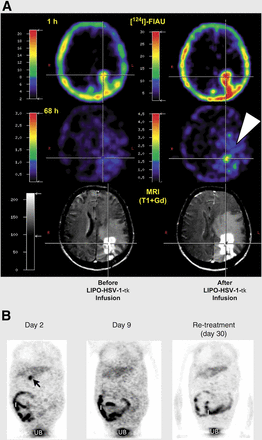
The promise of gene therapy and oncolytic viral therapy has yet to be realized, although both have been under investigation for decades. Monitoring the level of transgene expression or the distribution of oncolytic viruses would provide important, clinically relevant information that can be obtained only by noninvasive imaging. Preclinical animal studies have clearly shown that reporter imaging can track transgene expression and monitor oncolytic viruses (41). For example, HSV1-tk can be used as a therapeutic gene as well as a reporter gene. Although HSV1-tk has been used extensively in gene therapy clinical trials performed in the United States and Europe (42,43), only 3 clinical HSV1-tk gene imaging studies (Fig. 1) (36–38) and 1 hNIS gene imaging study (40) have been reported. Unfortunately, the pharmaceutical industry has been unwilling to image the magnitude of HSV1-tk expression in its clinical trials, even though the sensitivity of 124I-FIAU PET HSV1-tk expression has been shown to be more than 10-fold greater than that required for a therapeutic effect (Ronald G. Blasberg et al., unpublished data, April 1997).
Clinical studies. (A) HSV1-tk reporter gene imaging in patients after liposome-HSV-1-tk complex transduction. Shown is coregistration of 124I-FIAU-PET and MR imaging before (left column) and after (right column) HSV1-tk vector injection. A region of specific 124I-FIAU retention (at 68 h) within tumor is visualized (arrowhead). This tumor region showed signs of necrosis (cross hairs, right column) after ganciclovir treatment. However, only 1 of 5 patients showed 124I-FIAU–associated radioactivity localized at site of HSV1-tk vector injection. (Adapted from (36).) A similarly designed study, using 123I-FIAU and SPECT to monitor HSV1716 replication before and after administration of virus in 8 patients with high-grade gliomas, also failed to detect HSV1716-tk expression (37). (B) Adenoviral transgene (HSV1-tksr39) expression in patients with liver cancer. Coronal PET images 1.5 h after injection of 18F-FHBG were obtained 2 and 9 d after first dose of AdCMVtk and 2 d after readministration (day 30) of adenoviral vector in same lesion. Specific 18F-FHBG accumulation in treated tumor can be seen only at day 2 (arrow), and no specific accumulation can be seen either at day 9 or in retreatment. Radioactivity is observed in intestine (I) and urinary bladder (UB), as result of normal physiologic clearance of tracer. Late images obtained up to 6.5 h after injection did not show specific tracer accumulation in lesion either on day 9 or in retreatment study (images not shown). (Adapted from (38).)
Adoptive Therapies: Clinical Trials
A noninvasive method for repetitive evaluation of adoptively administered cells (e.g., bone marrow transplantation and immune cell–, stem cell–, and progenitor cell–based therapies) would benefit the assessment of current adoptive therapies, as well as future adoptive therapies using stem cells. The reporter systems that are being explored most intensively are adoptive therapy strategies to treat cancer (42) and heart disease (44). Individual patient monitoring would contribute to patient management by visualizing the trafficking, homing–targeting, and persistence of adoptively administered cells (using constitutive reporters) and would assess their functional and activation status (using inducible reporters, where the promoter–enhancer elements are regulated by specific endogenous transcription factors). For example, our group has monitored and assessed T-cell receptor–dependent activation in vivo using noninvasive PET (45). Such studies would significantly aid in the clinical implementation and management of new therapeutic approaches based on the adoptive transfer of immune cells, progenitor cells, and stem cells. Such imaging strategies have been widely applied in experimental animals, using both optical and radionuclide-based reporter imaging (8). Only a single clinical study has been reported, in which a patient with glioblastoma multiforme was infused with autologous cytolytic CD8 + T cells (CTLs) genetically engineered to express a tumor-targeting receptor and the HSV1-tk reporter gene (39). After tumor resection approximately 1 × 109 CTLs were infused into the operative site, and 18F-FHBG PET scans were able to detect the infused CTLs. Several other clinical trials are open, and others are being initiated. However, a limitation of such studies is the cost and required institutional infrastructure (Fig. 2).
Infrastructure to image T-lymphocyte trafficking, targeting, and activation. Therapeutic antitumor immunity depends on highly migratory T-cell populations capable of trafficking between lymphoid and tumor tissue sites as well as infiltration and activation within microanatomic structure of tumors. Stable genetic labeling of adoptively transferred cells with various reporter genes can be used to circumvent temporal limitations of ex vivo radiolabeling. Peripheral human T cells can be harvested from individual patients, transduced in GMP facility with vectors bearing both constitutive and activation-inducible reporter genes, and put through a round of selection and expansion. Then, these autologous genetically labeled tumor-specific T lymphocytes can be reintroduced into same donor patient. After administration of radiolabeled reporter-specific substrates, adoptively administered T cells can be monitored with respect to their trafficking through body, their targeting and persistence in tumor, and their activation and functional status in tumor. T-cell trafficking and activation can be monitored in vivo noninvasively and sequentially with PET on daily or weekly basis, depending on half-life of radionuclide used to label reporter substrate. Thus, reporter gene imaging allows for reliable assessments of adoptive immunotherapy through repetitive visualization of cellular trafficking, persistence, proliferation, and function at target site.
Imaging Oncogenesis and Signaling Pathways
Genetic mouse models including reporter genes allow examination of the consequence of a specific gene alteration on the formation of tumors in their normal tissue environment and the onset and temporal dynamics of signal pathway changes in real time using noninvasive imaging. A proof-of-principle study that used bioluminescence imaging to detect and measure K-Ras–dependent lung tumor genesis involved genetically altered mice (46). This approach is highly versatile and easily translatable to other mouse models of cancer. However, reporter gene imaging of oncogenesis and tumor development can be performed only on animal models, usually with low-cost optical systems in mouse models.
Other examples of imaging oncogenesis include platelet-derived growth factor (PDGF)–induced gliomas (47), the PSA-luc prostate cancer model (48), and the pVEGF-TSTA-luc reporter for a mammary tumorigenesis model (49). Numerous reporter systems have been developed to image activation of specific signaling pathways, including endogenous regulation of transcription (an endogenous transcription factor drives an inducible reporter) (45,50), posttranscriptional modulation of translation (51), and protein–protein interactions (52).
Imaging Drug Treatment
A strong rationale exists for assessing the effects of drugs that target or affect specific signaling pathways (e.g., PDGF/EGFR/HER2 signaling or Ras/Raf/MEK/Erk- and PKB/Akt/mTOR-mediated pathways) by noninvasive imaging. The development and use of reporter imaging (and direct imaging) paradigms to evaluate the efficacy of molecule-targeted therapies is being pursued by many groups. Specific drugs that are known (or thought) to target specific signaling pathways and downstream effectors can be assessed in reporter-expressing animal models or transgenic/oncogenic reporter animals. For example, the E2F-reporter mouse model was used in longitudinal preclinical studies to study the effects of treatment with a drug (PTK787/ZK222584) that inhibits the PDGF receptor (47). The E2F-reporter mouse model makes it possible to investigate the importance of PDGF-related signaling pathways in glioma maintenance and to image the effects of drug treatment. Other examples include the p53- and HSP90-sensitive reporter systems (50,53).
Translational Infrastructure: Good Manufacturing Practice (GMP) Facilities for Clinical Studies (Biologics and Radiopharmaceuticals)
GMPs for clinical cell engineering and cell-based therapy of patients requires an institutional infrastructure that conforms with city, state, and federal regulations. The operational structure for cell-based GMP and clinical activities include the following: ex vivo expansion and transduction of a patient’s cells; the production of ancillary cell lines and retroviral or lentiviral packaging cell clones with the creation of master cell banks; the production of viral stocks for subsequent cell transduction of cell-targeting, therapeutic, or reporter genes; and the production of clinical-grade plasmid DNA for vaccines and for generation of vector stocks. A schematic outline is shown in Figure 2. A quality control unit is essential to oversee several areas: first, the development, maintenance, and implementation of all standard operating procedures and batch records; second, the continual monitoring of the facility’s environment; third, the review and release of the results of assays performed on critical raw materials (e.g., culture media and sera; manufactured products such as viral stocks, packaging cells, and master cell banks; and expanded and genetically engineered patient cells, plasmid DNA, and vaccines); fourth, maintenance of the calibration programs of all pieces of equipment; and fifth, maintenance of personnel training files. The batch records and manufacture records should be independently reviewed by the institutional office of clinical research. For the preparation of radiopharmaceuticals for clinical trials and for the routine practice of medicine, the institutional infrastructure must conform with both local and federal regulations. In the United States, this conformance is governed by a combination of U.S. Pharmacopeia (USP) chapters (notably, USP <823> Radiopharmaceuticals for PET–Compounding and USP <797> Pharmaceutical Compounding–Sterile Preparations), USP drug monographs, Food and Drug Administration GMP regulations (title 21 of Code of Federal Regulations part 212 for radiopharmaceuticals), and the manufacturing procedures and quality assurance controls that are claimed in investigational new drug applications. For clinical trials of investigational radiopharmaceuticals, strict compliance with USP <823> is required. USP <823> refers extensively to USP <797>, which considers radiopharmaceuticals as low-risk-level compounded sterile preparations that should be compounded in a certified class 5 primary engineering control located in a class 8 or better air environment. A quality control area adjacent to the radiochemistry GMP facility is necessary to provide the required pre- and postrelease quality testing for each manufactured drug. Batch records and manufacture records should be independently reviewed by the institutional office of clinical research. Two additional mechanisms that facilitate the initiation of clinical studies are radioactive drug research committees and exploratory investigational new drug applications. Under title 21 of Code of Federal Regulations part 361.1, the Food and Drug Administration describes regulations that in effect delegate the oversight for radiopharmaceutical research to specially constituted institutional committees: radioactive drug research committees. Because the mechanism of these committees is not appropriate for first-in-humans studies, the exploratory investigational new drug has recently been developed as an alternative by the Food and Drug Administration. Under the exploratory investigational new drug, an investigator may combine early human studies at a phase 0, or microdose, level as a basis for getting important information that may provide drug developers with an early indication of whether drugs (and radiopharmaceuticals) are promising (or lack promise) for further development.
FUTURE
Our understanding of disease processes will continue to expand rapidly as molecular–genetic studies continue to explore the basis of disease development and progression. The era of molecular medicine has begun, and the benefits to individual patient care are expected to be realized soon. Six complementary human reporter systems (gene + probe pairs) have been proposed as candidates for future reporter gene imaging in patients. Importantly, these human genes are less likely to be immunogenic than are the reporter genes currently used in animals (e.g., viral thymidine kinases, luciferases, and fluorescent proteins). Once a complementary reporter pair (gene + probe) has been approved for human studies, the major regulatory focus will shift to the particular backbone and regulatory sequence of the reporter construct and to the vector used to target reporter transduction to specific cells or tissue, both ex vivo and in vivo. Ideal vectors for targeting specific organs or tissues (tumors) do not currently exist, although this is an active area of human gene therapy research. Although 3 of the 4 clinical studies were performed on patients with brain tumors, a good brain radiotracer–based reporter system has yet to be developed and validated, primarily because of limited reporter substrate penetration of the intact blood–brain barrier.
The development and validation of versatile and sensitive noninvasive assays that do not require tissue samples will be of considerable value. Molecular imaging, and noninvasive reporter gene imaging in particular, will make important contributions to monitoring molecular–genetic and cellular processes both in animal models and in human subjects and will be used for monitoring and optimizing new treatment strategies.
DISCLOSURE
This work was supported by NIH research grants 2P50CA086438-16, 1R01CA163980-01A1, and 5P01CA094060-12. Financial support was also provided by the Federal Ministry of Economy, Family, and Youth and the National Foundation for Research, Technology, and Development of Austria. No other potential conflict of interest relevant to this article was reported.
Footnotes
Published online Jan. 14, 2013.
- © 2013 by the Society of Nuclear Medicine and Molecular Imaging, Inc.
REFERENCES
- Received for publication July 31, 2012.
- Accepted for publication November 26, 2012.