Abstract
522
Objectives: Digital phantoms have played an important role in nuclear medicine imaging research, such as evaluating and optimizing image reconstruction and processing techniques. While the 4D extended cardiac-torso (XCAT) realistically models human anatomy, the creation of populations of phantoms remains a challenge: simple scaling of organs does not capture realistic anatomical variations within different patients. One method was proposed to create phantom variations by mapping phantom labels to patient CT segmentation via deformable image registration techniques. A major difficulty is that the patient CT segmentation is tedious to generate, thus practically limiting the number of variations. Here, we present a novel unsupervised convolutional neural network (ConvNet) based approach to directly register XCAT phantom with patient CT scans without prior training. We show that the generated phantoms are highly realistic and truly captured the patients’ anatomical variations.
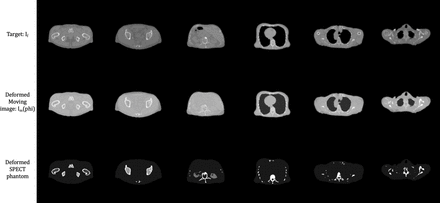
Methods: It is common to treat the image registration task as a variational problem where the total energy is equal to a regularization term plus an image similarity measure between the deformed moving and target images: E = R(phi) + Esim(Im(phi), If), where Im and If correspond to moving and target images, and phi represents the deformation field. In this work, we model the generation of the deformed moving image using a ConvNet based approach. It consists of two parts: a U-net like architecture to generate displacement field followed by a spatial transformer to deform moving image according to the produced displacements. The loss function of the proposed network is similar to the energy function, E, described previously. However, due to the large shape variations between the XCAT phantom and patients' CT scans, we did not require smoothness. Thus, the loss function was simply the image similarity measure, Esim(Im(phi), If); in this study, a Structural Similarity Index (SSIM) based similarity metric was used. Instead of following the traditional paradigm of training a neural network with a large set of training data, our proposed method is fully and truly unsupervised. The ConvNet optimizes the loss function for a given image pair iteratively via reparameterization in each iteration, independent from any prior training. Our model was implemented using Keras/Tensorflow on an NVIDIA Quadro P5000 GPU. We used 2D slices from a single XCAT phantom as moving images, and there were 1153 2D low-dose CT slices from 9 patients used as target images. SSIM and mean squared error (MSE) between the deformed moving images and the target images were computed to quantitively evaluate this method. SPECT simulations were also produced using the generated patient-like phantoms.
Results: We compared our method with a state-of-the-art registration algorithm, the symmetric image normalization (SyN) method. In terms of SSIM and MSE, the proposed method outperformed the SyN algorithm by a large margin, as shown in the table below. Qualitatively, SyN was able to capture the exterior variations within different CT scans, but it failed to model the details inside different organs and bone structures. By contrast, the proposed method successfully produced human-like phantoms with extremely realistic details.
Conclusions: We developed a fully unsupervised ConvNet based approach to generate patient-like phantoms that requires no prior training nor patient segmentations. Based on both quantitative and qualitative analysis, our method provided the best results compared to state-of-the-art conventional registration algorithms. Combined with simulation programs, the generated phantom can be easily transformed into realistic human-like nuclear medicine simulations.
Comparison of SSIM and MSE between the proposed method and the SyN algorithm.