Abstract
P524
Introduction: Due to the rapid tracer kinetics of Rubidium-82 (Rb-82) in cardiac dynamic PET, the significant cross-frame tracer distribution difference introduces substantial challenges to inter-frame motion estimation and correction, especially for the early frames, where traditional intensity-based image registration methods do not apply. We aim to develop a generative adversarial network (GAN) informed by temporal features to convert the early frames to the late reference frame for the purpose of improving the accuracy of frame-wise registration and parametric quantification.
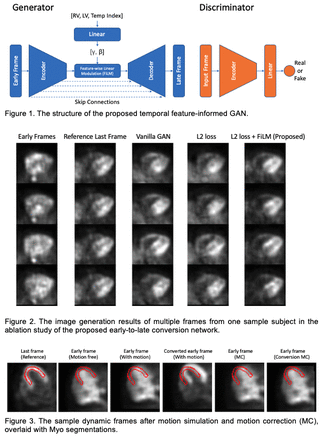
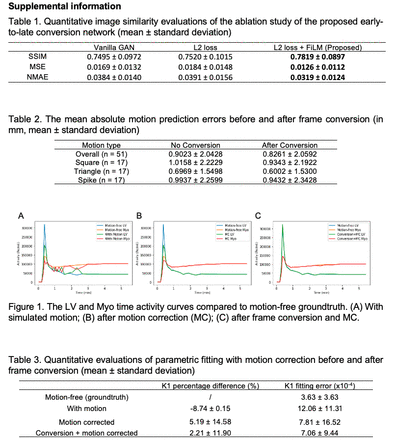
Methods: The study includes a total of 85 clinical Rb-82 PET scans (55 rest and 30 stress) from 59 patients that were acquired at Yale New Haven Hospital using a GE Discovery 690 PET/CT scanner and identified as motion-free by technologists. List-mode data of each scan in the first 6 min 10 s were rebinned into 27 dynamic frames. 17 scans were randomly selected for evaluation and the remaining 68 were for network training. The generator of the proposed GAN converts the input early frame to the corresponding late frame, employing the structure of a 3D U-Net with temporal feature-wise linear modulation (FiLM) at the bottleneck. A linear layer encodes the right ventricle (RV) and left ventricle (LV) blood pool time activity curves as well as the temporal frame index (Temp Index) as the auxiliary information and generates channel-wise parameters γ and β for the bottleneck FiLM manipulation. The discriminator of the proposed model encodes either the real or converted last frames and classifies them as either true or synthetic, using a PatchGAN architecture. The loss function consists of an adversarial loss and a voxel-wise L2 loss. The network is trained to convert all the early frames where the LV activity is equal to or higher than RV activity, to the last frame which was also the reference frame of the inter-frame registration. Qualitative visual comparison and quantitative image similarity metrics including structural similarity index measure (SSIM), mean squared error (MSE), and normalized mean absolute error (NMAE) were included for frame conversion evaluation. Following early-to-late frame conversion, three types (square, triangle, and spike) of simulated translational motion were added to both the original and converted frame sequences, resulting in 51 with-motion samples. Inter-frame motion correction was then implemented by mutual information-based registration implemented in BioImage Suite (BIS). The mean absolute motion prediction errors were calculated to measure motion estimation accuracy. After applying motion correction on the original motion-affected dynamic frames, we generated LV and myocardium (Myo) time activity curves and then fit the one-tissue compartment model for K1 quantification. The mean K1 percentage difference and fitting error were compared as the parametric fitting evaluation.
Results: The converted early frames had comparable visual quality to the reference last frames. With the newly introduced FiLM and L2 loss, the proposed network significantly improved SSIM, MSE, and NMAE compared with those of a vanilla GAN baseline. After frame conversion, the overall mean motion prediction error of BIS across all cases decreased from 0.90 ± 2.04 mm to 0.83 ± 2.06 mm. The LV and Myo time activity curves were closer to the groundtruth. In parametric quantification, the K1 percentage difference further decreased from 5.19% to 2.21%, and K1 fitting error further reduced from 7.81e-4 to 7.06e-4.
Conclusions: Temporally informed GAN could successfully perform early-to-late frame conversion for cardiac dynamic PET. The frame conversion could facilitate inter-frame motion correction with improved motion estimation accuracy and reduced parametric fitting error.