Abstract
P1360
Introduction: Lesion segmentation from PET images is paramount to treatment planning and for total metabolic tumor volume(TMTV) computation, which is relevant for patient outcome in lymphoma. Manual lesion segmentation is challenging as it is time-consuming and prone to errors due to intra- & inter-observer variabilities.
In this work, we train multiple 3D convolutional neural networks on cropped cubic patches of different sizes - as opposed to training directly on whole-body images-as a potential step towards improving segmentation performance. We also combine the predictions of these models via MajorityVote & STAPLE, to generate ensembled predictions.
Methods: Our lymphoma PET dataset(n=465) was annotated by 2 nuclear medicine physicians. We split the data into Train(n=298), Valid(n=75) & Test(n=93) sets. The PET images were resampled to (2,2,2) mm3 voxel spacing and the SUV values were normalized. Cubic patches of sizes (NxNxN), where N=96, 128, 192 & 256 were cropped from each 3D image with a probability of 4/5 for the patch being centered around a lesion.
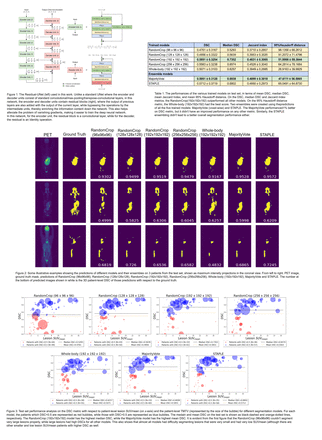
We trained four Residual-UNet models (Figure 1) for each value of N (RandomCrop(NxNxN)) using DiceLoss (data augmentation on patches; 1200 epochs). The validation was performed using a sliding-window method on whole-body images. As a baseline, we also trained a model directly on whole-body images (resized to 192x192x192) with the same preprocessing steps.
We aggregated the test predictions of each of the 5 trained models via two methods: voxel-wise MajorityVote & STAPLE. We evaluated the performance for all the models using Dice similarity coefficient (DSC), Jaccard index (JI) & 95% Hausdorff distance (95%HD) (in voxels).
Results: Among the 5 trained models, the N=192 model outperformed the other 4 on mean DSC, median DSC & JI metrics with values 0.585±0.325, 0.735 & 0.483±0.306, respectively; the baseline (Whole-body (192x192x192)) achieved values of 0.507±0.310, 0.626 & 0.395±0.268 respectively. It failed to be better than baseline on 95%HD (52.0±59.4 & 26.9±40.0, respectively). TheN=96 & 128 models had worse performance on all metrics (N=128 better than N=96) compared to both N=192 & baseline. The N=256 model also outperformed the baseline on DSC (0.556±0.324), median DSC (0.657) & JI (0.453±0.304), but not on 95%HD (64.3±78.2). Hence, N=192 model was the best on DSC and JI metrics, and the baseline was the best on 95%HD (Table 1, Figure 2).
We aggregated the predictions of all the models using MajorityVote & STAPLE methods. The MajorityVote obtained DSC, median DSC, JI & 95%HD of 0.595±0.313, 0.694, 0.490±0.302 & 47.6±56.6, respectively, while STAPLE obtained 0.571±0.372, 0.680, 0.465±0.297 & 65.0±64.9, respectively. So, compared to N=192, the MajorityVote was about 1% better on DSC, had a lower median DSC by 4%, a comparable JI & a better 95%HD by 4 (although it was worse than baseline on 95%HD by about 20).
We also visualized the test set DSC as a function of lesion SUVmean and TMTV. Figure 3 shows that while different models had different performances for segmenting each patient, almost all models had difficulty segmenting lesions that were very small & had very low SUVmean (although there are other smaller & low lesion SUVmean patients with higher DSCs as well).
Conclusions: In this work, we trained segmentation models with cubic patches of different sizes as well as a baseline on resized whole-body images. The best DSC and JI were achieved by the model with N=192, while the baseline had the best 95%HD. We hypothesize that for segmentation, there is a trade-off in segmentation performance between training with higher contextual information (large patch N=256 or whole-body) or focusing closely on the lesion (smaller patch N=96, 128). For the baseline, the whole-body images were also resized to 192x192x192, leading to a degradation in image quality. Further investigations are needed to validate this hypothesis more strongly, such as validation on different PET datasets and using different 3D neural nets.