Abstract
P1154
Introduction: Parkinson's disease (PD) is a neurodegenerative disorder which 5-10% of cases are only inherited in familial PD with mutations identified in several genes such as leucine-rich repeat kinase 2 (LRRK2), Synaptojanin 1 (SYNJ1) and others. Genetic issues lead to a 25% increase in the risk of PD. Prediction of pathogenic variants enables improved surgery of regions in the brain or symptomatic therapy. Semi-supervised machine learning approaches enable one to more readily increase training data sizes through usage of unlabeled data, in addition to existing labeled data. We aimed to investigate prediction of two important genes mutations, namely LRRK2 and SYNJ1, via semi-supervised hybrid machine learning systems (HMLS) using imaging and non-imaging data.
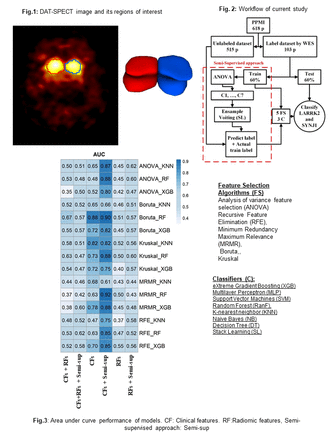
Methods: We collected 618 patients from the Parkinson's Progression Markers Initiative (PPMI) database, wherein LRRK2 and SYNJ1 gene statuses were considered as outcome. We first selected 103 patients out of the 618 patients utilizing their whole-exome sequencing (WES) as labeled dataset, while utilizing the rest of the patients (n=515) without labeled information (details of semi-supervised approach are below). Our labeled component included 32 patients with LRRK2 gene mutation, 24 patients with SYNJ1 gene mutation, and 47 patients who had other gene mutation types. Next, 55 clinical features (CFs) including motor and non-motor, as well as conventional imaging features (CIFs) from DAT SPECT images were extracted from the PPMI database. In addition, 215 radiomics features (RFs) were extracted from the SPECT images in the segmented regions of interest (left and right Striatum) using the standardized SERA software. We generated 3 datasets including CFs alone, RFs alone, and CFs+RFs. 60% and 40% of labeled patients were divided for training and testing purposes, named original labeled training and testing datasets. The datasets were normalized by the z-score technique. In semi-supervised approach, we employed 7 classifiers, namely Xtreme Gradient Boosting (XGB), Random Forest (RanF ), K-nearest neighbor (KNN) and others linked with analysis of variance feature selection (ANOVA) and grid search optimization algorithm, to classify the unlabeled datasets (515 patients). Thus, we used the original labeled training dataset (41 patients who had label) and the mentioned classifiers to predict class of the unlabeled patients. Meanwhile, Ensemble voting was applied to the predicted outcomes of these classifiers to determine the final class of the unlabeled patients. We employed 308, 246, 296 subjects out of 515 patients who had genes LRRK2 or SYNJ1 for CFs alone, RFs alone and CFs + RFs datasets, respectively. Overall, in our semi-supervised approach, we used the patients labeled by the classifiers in addition to original labeled training dataset to train the HMLS including 5 feature selection algorithms such as Minimum Redundancy Maximum Relevance (MRMR), ANOVA and others linked with 3 classifiers including KNN, RanF and XGB. Moreover, we employed original labeled training dataset alone to train HMLS. Finally, the original labeled testing dataset was utilized to evaluate our models.
Results: Semi-supervised approach linked with CFs alone + MRMR + RanF depicted the highest Area under curve (AUC) of 0.92, while the same HMLS with original labeled dataset had a lower AUC of 0.63. In RFs alone, semi-supervised approach + ANOVA + RanF arrived at an AUC of 0.60 while the same HMLS with original labeled training dataset achieved an AUC of 0.45. In datasets with both CFs and RFs, the highest AUC of 0.67 was provided by original labeled training dataset + Relief + RanF while the same HMLS linked with semi-supervised approached arrived at an AUC of 0.57.
Conclusions: This study showed that we can increase training data via our in-house approach semi-supervised which led to an improvement in prediction performance.