Abstract
2406
Introduction: With PET/CT, the CT images represent a measurement of photon attenuation in the subject and are used to accurately characterize the PET signal transmission through attenuation correction (AC). However, the PET and CT acquisitions are not performed simultaneously, and subject misalignment between the multi-modal image pair is often observed, leading to quantitative artefacts in the reconstructed PET images. Image registration can potentially address this spatial misalignment problem. However, established inter-modal approaches tend to be computationally expensive and challenging to automate – hence, they are typically not performed during image reconstruction.
This work presents a deep learning approach designed to optimize the CT image volumes for correcting the PET data based on inter-modal 3D elastic registration. It uses a convolutional neural network (CNN) to produce dense displacement vector fields (DVF), which is used to perform warping deformation at high resolution. The efficacy of the CT registration tool was evaluated in a test population of clinical PET/CT data for correcting spatial inconsistencies due to both physiological and bulk subject motion.
Methods: The CNN consists of a feature extractor and a DVF regressor. It takes as input a pair of uncorrected (NAC) PET/CT image volumes and estimates the DVF that characterizes their relative deformation field. Subsequently, this DVF is utilized to resample the CT image to match the PET. For the training, a supervised approach was devised within a dataset of well-aligned clinical PET/CT images – training samples were generated at each epoch using random patch selection and independent spatial augmentation. As such, each training input set comprised a pair of augmented PET/CT patch samples, with the deformation imposed between them serving as the “ground truth” target label. The entire network was trained end-to-end, and performance of the registration was continually monitored within the training and validation sets.
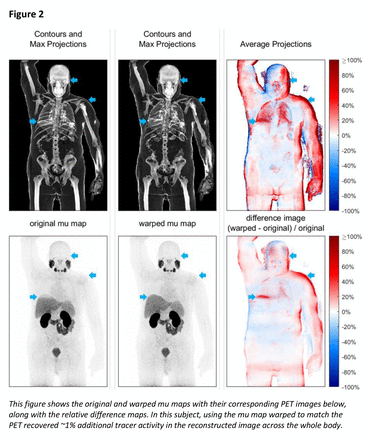
Real clinical data were used for testing, so there were no ground truth images of the PET tracer distributions. As such, the evaluation of this work is presented as a comparison with the default method, and so PET images were reconstructed both ways: with a warped mu map (wPET) and with the original, unmodified mu map (oPET). Images were compared, and qualitative findings are illustrated for different scenarios, including a specific focus on respiratory motion in whole-body and cardiac perfusion imaging.
Results: Matching the CT to the PET was found, in an independent population of test subjects, to reduce artefacts and improve tissue uniformity in the AC-reconstructed PET images. The qualitative behavior of the CT registration tool is illustrated for whole-body imaging using different PET tracers and also for physiologically gated cardiac PET data. Warping the CT produced PET images with generally higher quantified overall tracer activity - the mean activities measured within whole body test images were 67.3 MBq and 68.5 MBq for the oPET and wPET images, respectively. The paired t-test determined that these population differences were statistically significant (1.2±0.99 MBq, p = 0.002).
Conclusions: The PET/CT registration tool presented here was found to improve visual artefacts at various locations across the whole body in the validation datasets. Notably, this included “banana” respiratory effects occurring near the lung/liver border. Misalignment artefacts due to gross involuntary motion were also found to benefit from this approach. In particular, improvements were observed in cases where the subject adjusted his/her head position and for subjects which were scanned in the arms-up position and relaxed their arms throughout the PET acquisition. The feasibility of registering a single CT to every frame of a gated PET series is also demonstrated for cardiac imaging applications.