Abstract
1407
Objectives: Positron emission tomography (PET) with 18F-fluorodeoxyglucose (FDG) reveals metabolic abnormalities in Parkinson’s disease (PD) at a system level. Previous metabolic connectome analysis derived from groups of patients but do not identify individual neurophysiological details and the metabolic perturbations of basal ganglia and cortical connectivity that underlie the cardinal motor and non-motor features of PD. We present an individual metabolic connectome method to characterize the aberrant patterns of connectivity and topological alterations of the individual-level brain metabolic connectome and their diagnostic value in PD. Methods: FDG-PET data consisting of 52 PD patients, 62 healthy controls (HC) from our institution. Each individual’s metabolic brain network was ascertained using the Jensen-Shannon Divergence Similarity Estimation (JSSE) method. We analyzed the intergroup difference of the individual’s metabolic brain network and its global and local graph metrics to investigate the alterations of the metabolic connectome, including clustering coefficient (Cp), characteristic path length (Lp), normalized clustering coefficient (γ), normalized characteristic path length (λ), small-world (σ), global and local efficiency (Eglobal and Elocal), modularity score (Q), assortativity (Ar), hrierarchy (Hr) and synchronization (Sr). Also, the nodal property includes degree centrality, nodal efficiency, betweenness centrality, shortest path length and nodal clustering ecoefficiency. We used the multiple kernel support vector machine (MK-SVM) to identify the PD from HC individuals and conducted the nest leave-one-outcross-validation strategy to verify the performance of the methods.
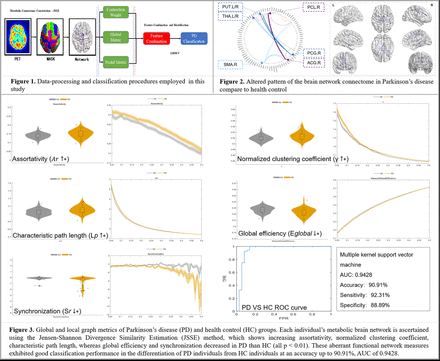
Results: The JSSE method showed the most involved metabolic motor networks were PUT-PCG, THA-PCG and SMA pathways in PD, which was similar to the typical group-level method, and yielded another detailed individual pathological connectivity in ACG-PCL, DCG-PHG and ACG pathways. PD individuals showed higher nodal topological properties (including Ar, λ and Lp) than HC individuals (all p < 0.01), whereas global efficiency and synchronization were lower in PD (both p < 0.01). These aberrant functional network measures exhibited good classification performance in the differentiation of PD individuals from HC individuals at an accuracy up to 90.91%. Conclusion: JSSE method identifies individual-level metabolic connectome of FDG-PET providing more dimensional and systematic mechanism insights for PD. The proposed classification method highlights the potential of individual’s connectome-based metrics for the identification of PD.
Global and local graph metrics of the brain connectome