Abstract
3005
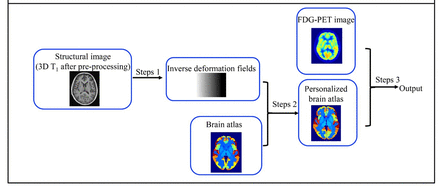
Objectives: Quantitative analysis of brain PET depends on structural segmentation, which can be time-consuming and operator-dependent when performed manually. Previous automatic segmentation methods based on an atlas usually fitted subjects’ images onto an atlas template for group analysis, which changed the individuals’ images and probably affected regional PET quantitation. We proposed an automatic segmentation method, registering atlas template to subjects’ images (RATSI), as shown in figure 1, which created an individual atlas template and may be more accurate and useful for PET quantitative analysis.
Methods: We segmented two representative brain areas in twenty Parkinson disease (PD) and eight multiple system atrophy (MSA) patients performed in hybrid positron-emission tomography/magnetic resonance imaging (PET/MR). The segmentation accuracy was evaluated using the Dice coefficient (DC) and Hausdorff distance (HD), and the standardized uptake value (SUV) measurements of these two automatic segmentation methods were compared, using manual segmentation as a reference. Further, RATSI was used to compare regional differences in cerebral metabolism pattern between PD and MSA patients.
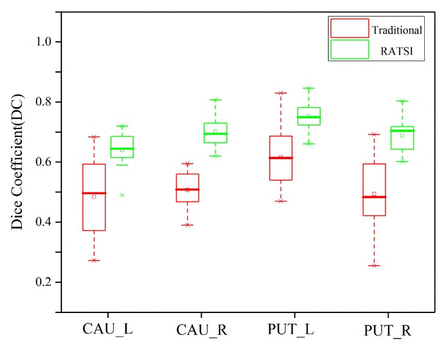
Results: The DC of RATSI increased and the HD decreased significantly (P < 0.05) compared with RSIAT in PD, while the results of one-way analysis of variance (ANOVA) found no significant differences in the SUVmean and SUVmax among the two automatic and the manual segmentation methods. For quantification in MSA and PD, the SUVmean in the segmented cerebellar gray matter for the MSA group was significantly lower compared with the PD group (P<0.05), as shown in figure 2, which is consistent with previous reports.
Conclusions: RATSI was more accurate for the caudate nucleus and putamen automatic segmentation, while has little effects for their max and mean SUV calculation in hybrid PET/MR.