PET, conventional nuclear imaging, and most contemporary medical imaging modalities are inherently digital technologies. Over the last several decades, there has been a transformative evolution of the digital computing landscape with respect to speed, cost of storage, infrastructure, and available expertise. However, our use of data, and in fact our whole understanding of the role of data in relation to emission imaging, has remained relatively unchanged. If we take a moment to reflect on this resource, generated ubiquitously in our daily imaging procedures, we can recognize that we have the capacity to support information use beyond the present convention and that the raw data provided by nuclear imaging studies can be tapped to fuel innovation.
Our general understanding of image data is that it exists in DICOM-format images, essentially analogous to film and representing a quantity of source signal distributed in space. However, the signals and information used to create these images in nuclear medicine originate in a much denser form; our imaging machines capture highly detailed time, location, and energy information for individual decay events. The current practice in PET, for example, is to truncate this information using assumptions and reconstruction techniques so as to provide a representation of tracer emissions distributed in recognizable Cartesian space. This process of biodistribution–representative image generation has essentially defined nuclear imaging for half a century. The procedure of truncating (unused) information is heavily ingrained in our practice likely because, for most of the field’s existence, it has been expensive and impractical to save raw acquisition data.
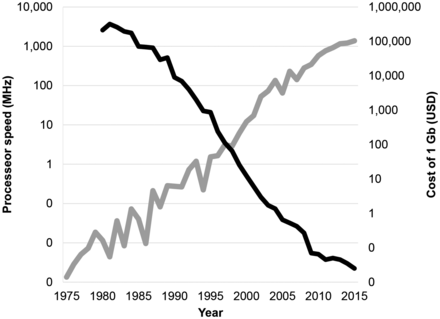
The costs associated with saving data have never been a static consideration. In 1980, a gigabyte of data cost $600,000 (1) (approximate value, inflation-adjusted), in 1990 that cost went down to $15,000, in 2016 it went down to $0.02, and we can confidently project continuation of this financial trend. Retaining a 2-Gb raw PET acquisition file now represents approximately 0.001% of the market cost of a scan. Both the cost and the capacity of digital imaging have undergone a slow but, in aggregate, very large shift. Each year, data-driven solutions become more practical and more relevant than in the year before, as shown in Figure 1. In the 1990s, we passed a milestone when digital storage became more cost-effective than paper storage (2). It is possible that we have now passed a new barrier in that we can say the cost of saving raw imaging acquisition data is negligible relative to the cost of generating the data. Furthermore, with ionizing radiation imaging, the cost-of-data paradigm does not include only an economic cost. Because patients are being exposed to radiation to generate these data, and at a risk to their health, it is prudent for us to periodically reconsider whether our practices are making optimal use of it.
PET is supported by a digital infrastructure that has undergone large transformations in the last few decades. Illustrated here are speed of processors (gray) and cost of storage (black) (1) shown sample-averaged across the years. All data are shown on log scales. Processor speed is extrapolated from a collection of historical transistor count references (11).
One reason to support changing our data-saving practice toward more robust access and archiving comes from the fact that we already have a body of literature showing that access to raw data can enable creative innovation. As an example, our group has recently published a study showing that large populations of scans can be corrected for motion using advanced data-use techniques and without the need for gating equipment or modified acquisition procedures (3). Additional areas of respiratory, cardiac, and head motion correction; signal and dose optimization; open-source reconstruction; and retrospective reframing have also begun to be explored (4). Progress in these areas and the impact of data-based innovation efforts have been limited because of the rigid data-access framework we currently have in place. We delineate acquisition data as proprietary, which subsequently impedes the academic and commercial exploration that traditionally propels potentially impactful ideas beyond the labs they are created in. For example, third party commercial or open-source standardized PET image reconstruction techniques could be developed in a clinically usable form and enable new levels of image quantification standardization across PET machines and centers and boost statistical power in research and multicenter clinical trials (5). Standardization can also make a large impact in the newly emerging fields of big data (6), machine learning (7), and radiomics (8). Arguably, there is no single effort that would more positively affect the latter two areas of research. Looking in another direction, we can also recognize personalized image reconstruction as a promising area of study, with efforts for task-based optimization emerging (9) and demonstrating the importance of researcher access to raw data. With respect to industrial innovation, accessible raw data would mean that those wishing to innovate its use commercially, such as to build a distributable data-driven gating (3,10) solution, would have access to raw data to develop the technologies, incentive to obtain regulatory approvals, and the ability to distribute solutions throughout the community. The third-party DICOM-based innovations we have seen developed provide an example of how access to data in combination with commercial and market forces can bring creative solutions into clinical use.
Another argument to support changing our data-saving practices is that our field benefits when it cultivates low-cost innovation. Lower costs generally mean greater inclusion of the research community for developing and benchmarking products and ultimately greater access for clinical end users. Looking forward, there is also an imperative to ensure our field continues to provide relevant leadership in the future. Attention to both the costs and the benefits of our innovations is important for ensuring our technologies provide relevant benefit in clinical care. In the United States and globally, we are witnessing efforts to combat the inflating costs of health care. Traditional strategies of investing in expensive, powerful technologies and waiting for cost effectiveness to catch up may not be as viable a path for innovation as it has been in years past. Data-driven innovation stands in contrast to a hardware-driven model, as the former is centered on a concept of doing more with what you already have. What’s more, the trend of digital evolution not only empowers us to develop innovative uses of data but also assures us that if we tie our solutions to data-use technologies we can reasonably expect them to become faster, cheaper, and more powerful with time. Finally, nuclear medicine is a global field, and recognizing that data produced in all our systems are a valuable tool for innovation would enable inclusive paths toward innovation that expand our pool of creative talent and potential leaders.
The final rationale we present to support changes in our data-saving practices addresses what we do not know about the future. Even if there were an insufficient argument for changing our practice today, we can look forward and see how changes now may benefit the pioneers of tomorrow. What if, for example, we were better stewards of medical data in the 1990s? We would now be decades ahead in our efforts to mine and interpret big data. We are now changing our practices to archive electronic medical records and image data, but we are not commonly archiving raw data. We admittedly do not know what the future will bring. However, recognizing now the potential value of raw data, identifying it as a resource, and preparing it to be harvested by future generations is within our present capacity. It is not difficult to imagine how very near-future innovations in radiomics and computer-aided diagnosis would benefit greatly from access to raw data along with archived medical records for benchmarking new techniques in large, standardized, associative studies. Looking forward, we can also consider that the newly trained and future generations of imaging professionals, as well as the patients they are serving, will be digital natives and will likely have talents and expectations for data management that go beyond the current standard.
The benefit of data valuation extends across the medical imaging fields. However, the nuclear medicine community is favorably positioned to play a leading role in redefining the value of raw acquisition image data. Our data are inherently filled with useful timing, energy, and spatial information. Nuclear imaging is used for a variety of applications that span the medical specialties. Our field has long included cooperation among a variety of specialists within our community: physicians, physicists, computer scientists, mathematicians, and others. This diversity and history of cross-specialty collaboration places our field in a favorable position to pioneer new concepts on the value of data and advanced data-use–based innovation.
In an effort to coalesce the ideas mentioned here, we take this opportunity to present the concept of small data for the nuclear medicine and imaging communities: Small data are defined as informative, possibly ancillary details inherent within data or datasets. Small data are local, actionable, often personalized elements of information that can inform and enable optimal utility. The term small data encompasses the notion that every digital bit of information may have value and utility and implicitly implies the importance of its access.
Currently, there is much excitement about using the enormously increased power of computer systems to mine big data, that is, data that are stored across multiple databases and capture information from various sources, such as genetics, treatments, and long-term patient outcome. We argue that modern computing power should also be applied to small data, the raw data that are routinely acquired every day during clinical practice.
The term small data in not entirely new. At present, various mentions can be found on the Internet. To our knowledge, however, it has not been defined as a concept of data valuation formally or in peer-reviewed publications. We are taking this opportunity to do that and to clarify its relevance to the imaging community. Small-data innovation encompasses technologies that use small-data details and represents an area with potential for meaningful imaging innovation. Small-data innovation may include revisiting traditional uses of data and extracting greater details or using modern computing power to develop new or improved processing strategies. Small-data innovation can support personalized imaging or task-optimized imaging through advanced information extraction techniques, personalized image processing, and dose optimization.
In the 20th century, a main challenge of nuclear medicine and radiology information technology was reconstructing 3-dimensional images and displaying and storing the reconstructed images efficiently. As we progress into the 21st century, the vastly increasing processing power, network bandwidth, and storage capacity of current computer systems now allow us to go beyond the reconstruction of the distribution of radioactivity at a given time. By storing and analyzing the raw image data, we can derive additional information, such as signal from motion, spatial or temporal characterization of signal dependability, or quantitative uptake measurements based on open-source standardized reconstruction strategies. By reviewing and updating data access practices, we can open the door to new, clinically applicable commercial innovations in the same manner as that by which we found success with the DICOM image standardization initiative of the 1990s. What is required now is not expensive investment but rather an open mind in our community toward exploring the potential benefits of understanding and using small data differently.
DISCLOSURE
No potential conflict of interest relevant to this article was reported.
Footnotes
Published online Nov. 22, 2016.
- © 2017 by the Society of Nuclear Medicine and Molecular Imaging.
REFERENCES
- Received for publication July 15, 2016.
- Accepted for publication November 10, 2016.