Abstract
242428
Introduction: Myocardial perfusion imaging (MPI) is widely used non-invasive technique for assessment of myocardial blood flow (MBF) and guiding treatment of patients with coronary artery disease (CAD). Traditionally, MPI is interpreted based on static images and semi-quantitative scores. Quantitative parametric imaging using traditional nonlinear least squares (NLS) fitting method faces challenges such as limited coincidences, complex and time-consuming computation, etc., while deep learning (DL) models are unavailable since lacking of ground truth parametric images (PIMs) for training. This study aims to utilize virtual clinical trials (VCTs) to assess a deep neural network's effectiveness in parametric imaging for MBF estimation.
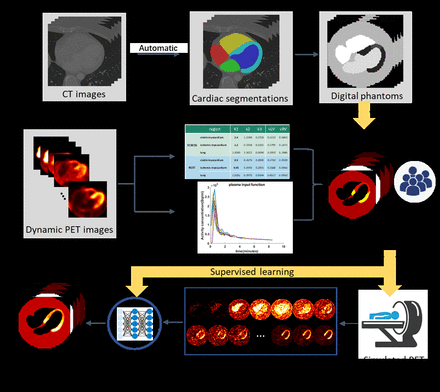
Methods: The main steps of VCTs involve generating digital phantoms and a virtual population, simulating an imaging system, and interpreting the model using the known ground truth data. In this study, digital phantoms utilized cardiac segmentations obtained from 95 cases of clinical CT images. These segmentations consisted of 5 regions: left ventricular (LV) myocardium, left chamber, right chamber, a circular background, and a random 3D volume added to the LV myocardium to simulate ischemic defect. Ten patients who underwent both rest and stress [13-N] ammonia PET/CT scans were used to derive averaged arterial input function (IF) and kinetic parameters (K1, k2, k3, vLV, vRV) for the irreversible 2-tissue compartmental model in virtual population simulation. To incorporate biological variability, a 30% variation was introduced to the first 10 time points of both rest and stress IFs, and they were further shifted either left or right by one frame step. The noise-free PIMs and IFs were used to synthesize pristine dynamic PET images. Analytic simulation of projection data as well as OSEM reconstruction were conducted. With each phantom assigned 4 distinct kinetic parameter settings by sampling these parameters from a uniform distribution, a total of 440 cases (rest/stress 55x4 = 220 cases) were generated for network training. An improved transformer network was adopted in this study. The network takes 21-frame 4D PET images as input and outputs 5 PIMs. We trained a rest model and a stress model separately using 5-fold cross-validation with 200 epochs. We evaluated the performance of the DL model on the external testing dataset (rest/stress: 40x4 = 160 cases) through the mean absolute relative error (MARE) calculated on the myocardium.
Results: For the parameter of interest, the stress K1 of the viable myocardium ranged from 1.2 to 3.6 (mL/(g*min), and the rest K1 ranged from 0.6 to 1.2 in the data simulation. For both rest and stress study, upon visual comparisons to the traditional NLS method, the images obtained by our DL method show a notable reduction in noise and clearer tissue structure. For the rest model, quantitative analysis showed that the DL method significantly improved the MBF estimation from the MARE of 47% (NLS) to 23%(DL) (p<1e-10). As K1 values decreased from 1.2 to 0.6, the bias increased by about 20% (NLS: 21%, DL: 18%). For the stress data, the MARE of the myocardium K1 estimated by the DL model was significantly lower than the NLS method (NLS: 51% vs. DL: 25%, p<1e-10). As stress K1 decreased from 3.6 to 1.2, the estimation bias was considerably increased by 35% for the NLS method, and 20% for the DL method.
Conclusions: The results demonstrated the feasibility and effectiveness of a VCT-based DL model in parametric imaging of MBF PET. This work shows how VCTs enable study of essential factors regarding the MBF quantification in the context of lacking of ground truth measurements of MBF, and can avoid costly and risky clinical trials.