Abstract
241979
Introduction: Cancer is a worldwide health concern and the second leading cause of death in the United States. Automatic detection and characterization of cancer are important clinical needs to enable early treatment. We developed a deep transfer learning approach for fully automated, whole-body tumor segmentation and prognosis on positron emission tomography (PET)/computed tomography (CT).
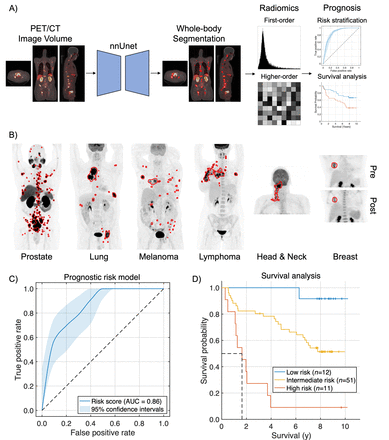
Methods: De-identified data from 611 2-deoxy-2-[18F]fluoro-D-glucose (FDG) PET/CT scans of patients with lung cancer, melanoma, lymphoma, head and neck cancer, and breast cancer and 408 prostate-specific membrane antigen (PSMA) PET/CT scans of patients with prostate cancer were used in this study. A subset of the data was collected, in part, from The Cancer Imaging Archive. The approach jointly optimized a 3D nnU-net backbone, an automatically self-configuring DL framework, and learned the generalized segmentation task across the source and target domains of FDG and PSMA PET/CT images, respectively, using deep transfer learning (Fig. 1a). Limited manual annotations were used for training. True positive rate (TPR), positive predictive value (PPV), Dice similarity coefficient (DSC), false discovery rate (FDR), true negative rate (TNR), and negative predictive value (NPV) were assessed to evaluate segmentation performance. Radiomic features and imaging measures quantifying molecular tumor burden and uptake were extracted from the predicted segmentations. Prognostic models were developed using the extracted imaging measures to perform risk stratification of prostate cancer based on follow-up prostate-specific antigen (PSA) levels and PSA doubling times, survival estimation of head and neck cancer by the Kaplan-Meier method, and Cox regression analysis, and prediction of pathological complete response (pCR) for patients with breast cancer undergoing neoadjuvant chemotherapy. Overall accuracy and area under the receiver operating characteristic (AUROC) curve were assessed.
Results: Illustrative examples of the predicted tumor segmentations are shown in Fig. 1b. Our approach yielded median TPRs of 0.75, 0.85, 0.87, and 0.75, median PPVs of 0.92, 0.76, 0.87, and 0.76, median DSC values of 0.81, 0.76, 0.83, and 0.73, and median FDRs of 0.08, 0.24, 0.13, and 0.24 for patients with lung cancer, melanoma, lymphoma, and prostate cancer, respectively, on the voxel-wise tumor segmentation task. The approach yielded median TNRs and NPVs of 1.00 across all patients. The prognostic risk model for prostate cancer yielded an overall accuracy of 0.83 and an AUROC of 0.86 (Fig 1c). Patients classified as low-, intermediate- and high-risk had mean follow-up PSA levels of 9.18, 26.92, and 727.46 ng/mL and mean PSA doubling times of 8.67, 8.18, and 4.81 months, respectively. Patients with head and neck cancer were stratified based on the risk score and plotted using Kaplan-Meier estimators (Fig. 1d). Patients predicted as high-risk had a shorter median overall survival compared to low- or intermediate-risk patients (1.64 years vs median not reached, P < 0.001). Patients predicted as intermediate-risk had a shorter median overall survival compared to low-risk patients (P < 0.05). The risk score for head and neck cancer was significantly associated with overall survival by univariable and multivariable Cox regression analyses (P < 0.05). Predictive models for breast cancer predicted pCR using only pre-therapy imaging measures and both pre-therapy and post-therapy measures with overall accuracies of 0.72 and 0.84 and AUROC values of 0.72 and 0.76, respectively.
Conclusions: A deep transfer learning approach was developed for fully automated whole-body PET/CT tumor segmentation and generalized across patients with six different cancer types imaged with FDG and PSMA-targeted radiotracers. The approach automatically quantified imaging measures of molecular tumor burden and demonstrated prognostic value for risk stratification, survival estimation, and prediction of treatment response.