Abstract
241482
Introduction: Alzheimer's disease (AD) exhibits an increased accumulation of extracellular amyloid-β (Aβ) plaques in the brain, which is associated with accumulation of intraneuronal neurofibrillary tangles built by tau-protein. Aβ and tau can be imaged non-invasively using [11C]-PIB (PIB) and [18F]-MK6240 (MK) through PET. Combined information from the two aforementioned hallmarks of AD synergistically can allow a more detailed understanding of their interaction, of the prodromal phase and the disease progression. At present, due to the intrinsic PET characteristics, the two tracers are imaged in separate sessions that are sometimes days apart, which jeopardizes the clinical feasibility and accuracy of multi-parametric studies. Dual-tracer PET offers natively aligned spatiotemporal information without requiring co-registration of two separate image sets. Furthermore, it can facilitate the clinical implementation of multi-parametric imaging and overcomes the logistical, cost, and throughput challenges of separate-day imaging protocols. Here, we assess the accuracy of the individual PIB and MK parametric maps when recovered from dual-[PIB+MK] dataset acquired with staggered radiotracers injections.
Methods: In a separate study, 20 cognitively healthy subjects from two different cohorts underwent 70min and 120 min dynamic PIB (527±23 MBq) and MK (190±15 MBq) PET, respectively on a Siemens BiographTM mCT PET/CT. The Logan distribution volume ratio (DVRs) reproducibility for each tracer were assessed with shorter dynamic scan times. Both PIB and MK DVRs were reproduced using only 40min of dynamic data, which has been adopted in what follows as the delay time between PIB and MK injections in a dual-tracer acquisition setting.
Specifically, four cognitively healthy subjects (2 male, 2 female, 67±3 yo) and 1 MCI/AD male (56 yo) underwent 80 min PIB and 40 min MK dynamic PET in two separate days.
PIB and MK PET dynamic images were un-decay-corrected and spatially co-registered on a frame-by-frame basis. First 40min of the MK dynamic PET was combined with the 80min PIB dynamic scan starting at 40min post-PIB injection to synthesize dual-[PIB+MK] PET scans with staggered injections. This resulted in an 80min dual-[PIB+MK] PET dynamic dataset.
Two-tissue compartment kinetic modeling (k4 ≠ 0) of the dual-[PIB+MK] dynamic PET was performed a on a pixel-by-pixel basis for the whole brain using the mfEVolveTM software. The individual tracer input functions were derived from the carotid arteries using the dynamic PET images. The resulting kinetic parameters were used to separate the two tracers signals from the dual-tracer dynamic imageset and then kinetic analysis was carried out for each separately using the Logan reference model with PMOD® 3.9. Finally, the corresponding Logan DVR parametric maps were compared to those deduced from the original separate tracer dynamic datasets (gold standard).
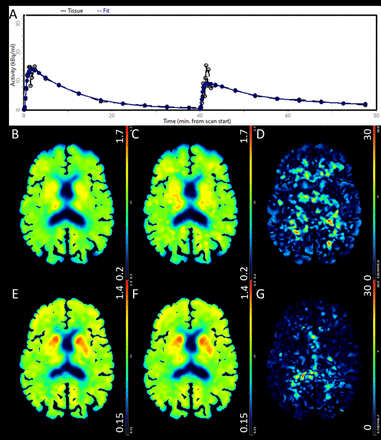
Results: Figure 1-(a) shows an example of the gray matter time activity curve fit using dual-[PIB+MK] compartmental kinetic modeling. The DVR parametric maps from the original and dual-tracer PIB imagesets as well as their corresponding percent differences are shown in figures 1- b, c, & d respectively. The corresponding images for the MK are presented in figures 1- e, f, and g. The brainstem and Accumbens nuclei DVR’s deduced from the dual-[PIB+MK] dataset exhibited the largest percent difference for PIB (6.50±1.96)% and MK (1.45±1.49)% respectively when compared to those derived from single-tracer dynamic datasets.
Conclusions: Compartmental kinetic modeling of dual-[PIB+MK] dynamic PET can reproduce the quantitative measures performed with standard single-tracer PET imaging. This can facilitate the clinical implementation of multi-parametric PET imaging in AD studies.