Abstract
241413
Introduction: The whole-brain segmentation based on integrated PET/MRI systems significantly contributes to both functional and anatomical quantification of the brain, offering precise diagnostic information for diverse brain disorders. Nowadays, the majority of whole-brain segmentation studies have been used only with MRI images, ignoring the importance of multimodal information fusion, which enable to greatly enrich the features during network training and improves the speed and accuracy of segmentation of the brain. Therefore, we aimed to employ deep learning techniques to achieve automatic and accurate segmentation of the whole brain while incorporating functional and anatomical information.
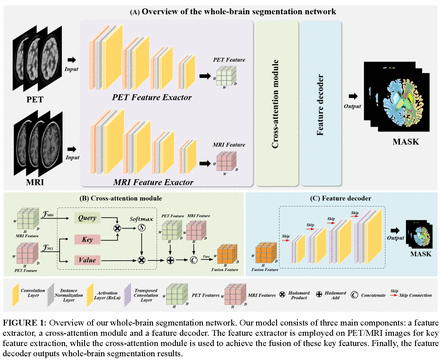
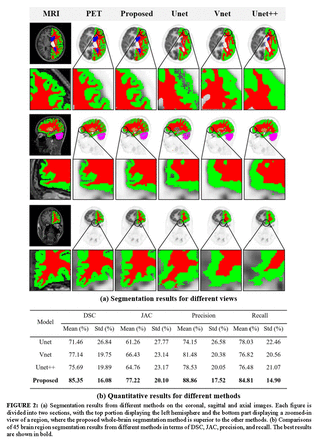
Methods: Experiments was conducted on 120 brain scans acquired from the integrated PET/MRI system. FreeSurfer software was used to register the PET/MRI image pairs and generate 45 brain mask data, which served as the ground truth. To leverage dual-modality information, we proposed a novel 3D network with a cross-attention module to capture the correlation between dual-modality features and improve segmentation accuracy. Moreover, several deep learning methods (Unet, Unet++, Vnet) were employed as comparison measures to evaluate the model performance, with the Dice similarity coefficient (DSC), Jaccard index (JAC), recall, and precision serving as quantitative metrics. Furthermore, correlation and consistency analyses based on different brain regions were also performed.
Results: Our model introduces the cross-attention mechanism for whole-brain segmentation with PET/MRI dual-modality data participated. Both the quantitative and visualization results demonstrated the advantages of our method in whole-brain segmentation. Compared with other deep learning approaches, our method obtained better model performance with 85.34% DSC, 77.22% JAC, 88.87% recall and 84.81% precision. In addition, violin plots showed that the SUV distribution of our method within the brain regions was consistent with that of the ground truth. Moreover, the chord plots also indicated that the correlation analysis based on our segmentation results could accurately reflect the relationships among different brain regions.
Conclusions: In this study, we have proposed a novel 3D network with a cross-attention module to achieve automatic segmentation of whole-brain structures, achieving outstanding segmentation accuracy. Consistent and correlated analyses also illustrated that our approach enable to achieve superior performance. Our method has shown great potential in personalized assessments and brain disease analyses with high efficiency and accuracy, and we will try to apply our method to other multimodal tasks, such as PET/CT data analysis, in clinical practice.