Abstract
P1609
Introduction: Alzheimer's disease (AD) is the most common pathological substrate for dementia, contributing to 60–70% of all dementia cases. Amyloid and tau proteins are the hallmarks of AD pathology. Positron emission tomography (PET) tracers that image amyloid and tau non-invasively are available and collecting both types of scans is useful for evaluating novel treatments and for studying the natural course of AD progression. However, tau PET is extremely expensive and not universally available, leading to great interest in estimating tau PET images from cheaper and more accessible measurements such as amyloid PET. In this study, we proposed a novel application of the Conditional Generative Adversarial Network (cGAN) framework to generate high quality tau PET (18F-flortaucipir) scans from the corresponding amyloid PET (18F-florbetapir) scans. To the best of our knowledge, this is the first study to estimate synthetic 18F-flortaucipir PET from 18F-flortaucipir PET.
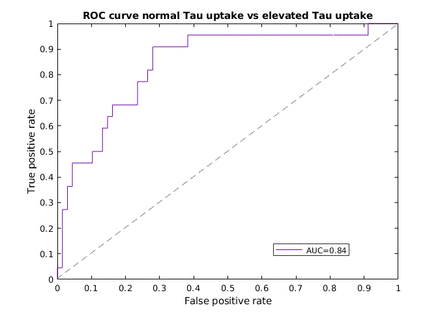
Methods: The data used in this study was obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) online depository. A dataset of 475 individuals who underwent both 18F-florbetapir and 18F-flortaucipir scans within a 6-month period with a clinical diagnosis of cognitive normal (n=231), mild cognitive impairment (MCI, n=73), early MCI (n=71), late MCI (n=33), subjective memory complaints (n=46), and AD (n=21) were included in our study. Fully preprocessed amyloid-tau PET pair images were normalized to the MCALT template and skull stripped, using SPM12. Due to the unbalanced nature of the dataset data augmentation was performed using x and y translation and flipping. The data was divided into training, validation, and test sets with a 70%/10%/20% split, respectively. cGAN models consist of two competing networks, a generator that creates synthetic data and a discriminator that tries to distinguish between real and synthetic data. Our cGAN model was trained using real, corresponding amyloid-tau PET pairs. The resulting model takes real amyloid PET images as input and outputs synthetic tau PET images. The developed cGAN was trained for 50 epochs with a batch size of 32 using a mean absolute error loss function. The generator is a U-net based convolutional neural network with skip connections. The discriminator is a convolutional Markovian discriminator. The similarity between synthetic and real tau PET images was evaluated via the structural similarity index (SSIM), peak signal to noise ratio (PSNR), and normalized root mean squared error (NRMSE). Furthermore, we extracted regional standard uptake value ratios (SUVRs) and meta-region of interest (ROI) SUVRs from the real tau PET scans and synthetic tau PET scans to quantify the model's performance. The meta-ROI have previously been shown to have wide dynamic ranges in normal aging, pathological aging, and AD dementia [1]. The utility of the synthetic tau PET meta-ROI SUVR for identifying real tau PET scans that are tau positive (defined in terms of a widely utilized definition of tau PET positivity) was evaluated in terms of true positive rate (TPR), false positive rate (FPR), and the area under the receiver operating characteristic (ROC) curve (AUC).
Results: Our cGAN model yielded a SSIM of 0.917, a PSNR of 27.04, and a NRSME of 0.18 on the test set. The ROC curve analysis (Figure 1) resulted in an AUC of 0.84%. By applying a cut-off threshold corresponding to the model optimal operating point, a TPR of 90% and FPR of 28% were achieved. The SUVR analysis yielded a mean absolute percentage error of 8.3% ± 6.5% on the test dataset. These results are competitive with other medical image generation using deep learning approaches [2-4].
Conclusions: The proposed cGAN model generated high quality 18F-flortaucipir PET images from 18F-florbetapir PET images without any anatomical information from MRI or CT. Its amyloid-based estimates of tau could be useful in a screening paradigm to use amyloid PET to identify those individuals most likely to be tau positive.