Abstract
P1585
Introduction: Deep learning methods, based on generative adversarial networks (GAN), have shown great potential in synthesizing medical images or transferring context between different medical imaging modalities. While it is premature to consider entire replacement of a modality with another (especially if the imaging principles are very distinct), it is desirable to quantify how truly complementary two modalities are. We believe that deep learning-based image-to-image translation can play a role to this end, since two modalities may be entirely distinct in their original scales (e.g. PET and CT). As demonstration, we investigate the relative complementarity of 18F-FDG PET and CT imaging of head and neck cancer, by synthesizing 3D-PET images from CT images, followed by comparison with real 3D-PET images.
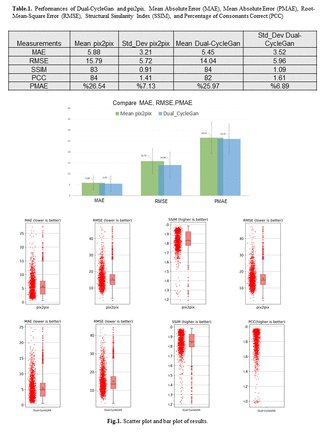
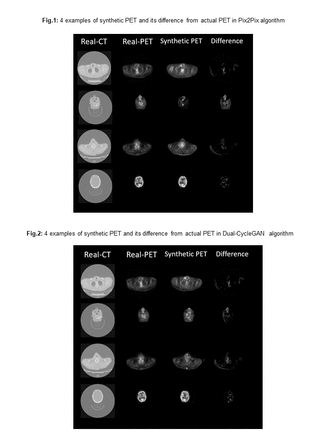
Methods: We studied a large dataset of 865 head and neck cancer patients with PET and CT images including size of 167x167x129. The datapoints were collected form The Cancer Imaging Archive database. PET images were first resampled 00ae_registeredsign to CT, and then both images were enhanced and normalized. We utilized Pix2pix and Dual-Cycle Generative Adversarial Network (CycleGAN) for image-to-image translations between domains. Pix2pix works in a pairwise fashion in that it needs corresponding images from two different domains to learn to translate from one to the other whilst CycleGANs need no corresponding images from both domains to learn. It learns a two-way mapping between the domains. In current study, we applied Pix2Pix and CycleGAN to synthesize 3D PET images from 3D CT images. The Pix2Pix parameters were experimentally tuned using L1 loss function, Adam optimizer with beta of 0.5 and learning rate of 0.0002. Moreover, the Dual-CycleGAN parameters were experimentally optimized using six independent loss terms to balance quantitative/qualitative loss functions including adversarial, dual cycle-consistent, voxel-wise, gradient difference, perceptual, and structural similarity losses, all with default initialization parameters. A novelty of this study is the usage of 3D images for training, instead of 2D images, and both networks were trained for 4000 epochs with a batch size of 8. The Mean Absolute Error (MAE), Percent Mean Absolute Error (PMAE), Root-Mean-Square Error (RMSE), Structural Similarity Index (SSIM), and Percentage of Consonants Correct (PCC) metrics were utilized to evaluate and compare the models. The datasets were split into 679 and 186 subsets for training and testing propose, respectively.
Results: The Dual-CycleGAN model had a MAE of 5.45±3.52, RMSE of 14.9±6.0, SSIM of 0.84±0.10, and PCC of 0.82±0.16. Moreover, the Pix2Pix model had a MAE of 5.88±3.65, RMSE of 15.8±5.7, SSIM of 0.83±0.09, and PCC of 0.84±0.14. Specifically, PMAE between synesthetic PET and true PET were (26.0±6.9)% and (26.5±7.1)% for Dual-CycleGAN and Pix2Pix, respectively. This indicates that there is significant information from PET that can be recovered from CT, yet the complementary value of PET to CT is significant at around 25%, though usage of (1-SSIM) suggests a complementary value of around 16%. This problem can be framed, ultimately, in terms of a task of interest with its specific metrics, which can be applied in this proposed framework for complementarity assessments.
Conclusions: Our study indicates that optimized Dual-CycleGAN and Pix2pix models generated considerable information from CT images in synthetic PET images. Meanwhile, the complementarity of PET and CT images was captures by around 25% between the synthesized and real PET images.