Abstract
P1451
Introduction: A Pre-trained deep neural network named "DnCNN" for image denoising is available with MATLAB Deep Learning Toolbox. This network is trained with natural images (such as images of tree, animals, buildings, and so on) taken with photographic camera. Such images have good photon statistics and may not be impacted if some details are lost during the process of denoising. In contrast, nuclear medicine images have very low photon statistics and loss of details during the denoising process will have serious consequences since the denoised image is used for diagnosis and treatment response evaluation.
In order to answer the question of whether this pre-trained DnCNN can be used without training on nuclear medicine images, a study is required. The time, energy, and financial cost involved in the designing and training a network from scratch could be eliminated if the pre-trained DnCNN could be used for denoising Tc-99m-DMSA images. Present study investigates precisely in this direction.
Methods: Two hundred forty-two images were denoised using the pretrained DnCNN network. The images were acquired on Siemens Symbia T6 SPECT CT dual head gamma camera fitted with LEHR collimator, 3 hours after administration of 185 MBq Tc-99m DMSA. The denoised image was compared visually with its corresponding input image by two nuclear medicine physicians. They especially looked for loss of clinical information if any in the denoised image that can have serious consequences in diagnosis or in treatment response evaluation.
The quality of denoised image was also assessed objectively using the image quality metrics PIQE, Blur, GCF, and Brightness. Wilcoxon signed rank test with continuity correction was applied to find the statistically significant difference between the value of image quality metrics of the denoised images and the corresponding original images at the level of significance = 0.05.
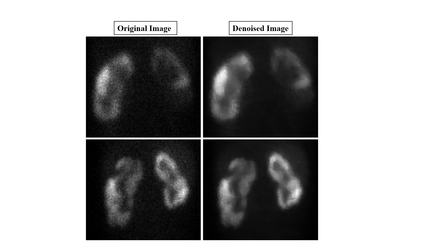
Results: Nuclear medicine physicians observed no loss of clinical information in denoised image compared to its input image [Figure 1]. The quality of denoised image was found to be superior compared to its input image both visually and objectively. Edges/boundaries of the scar were found to be well preserved, and doubtful scar became obvious in the denoised image [Figure 2]. Objective assessment (PIQE, Blur, GCF, and Brightness) also showed that the quality of denoised images was significantly better than that of original images at p-value <0.0001.
Conclusions: The pre-trained DnCNN available with MATLAB Deep Learning Toolbox can be used for denoising Tc-99m-DMSA images.