Abstract
P1193
Introduction: Prostate cancer treatment often relies on highly localized and targeted medical procedures, such as radiation therapy, biopsies, or excisions. Prostate-specific membrane antigen (PSMA) PET imaging, combined with CT imaging, facilitates such treatments. The combination of the two modalities provides both functional and anatomical information and has become a common imaging technique for patients with prostate cancer. High prostate lesion uptake in the PET scan results in relatively low-noise images of the pelvis. CT scans, on the other hand, provide high quality images for discerning organs such as the prostate but lack soft tissue contrast making accurate delineation more challenging. Accurate and reliable segmentation of the prostate would improve targeted interventions, however manual segmentation by trained physicians is expensive and introduces variability. Automating this process would not only help standardize individual treatment procedures, but also facilitate studies on the progression of the disease. We present a fully automated method of segmenting the prostate from PSMA PET-CT scans. We believe reliable prostate segmentation is a crucial step in developing accurate tools for standardized prostate lesion reporting.
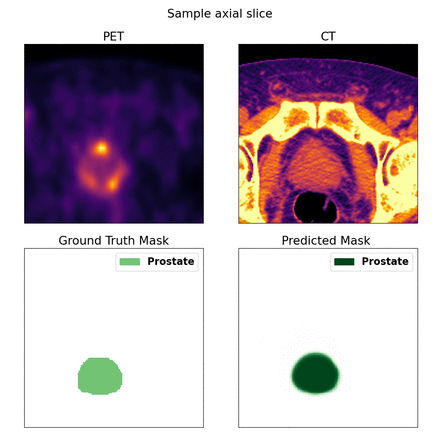
Methods: We propose a fully automated deep segmentation model for the prostate, using PSMA PET-CT imaging. For our dataset, 59 such scans were collected and labeled by a nuclear medicine physician using CT. An additional set of segmentations were created using a semi-automated thresholding method operating as the baseline approach. These 'threshold' segmentations were provided for a subset of the 59 patients (n=53). For model training, we used a 3-fold procedure: training set (n=40), validation set for model selection (n=9), and test set for evaluation (n=10). This dataset was used to train a 3D U-Net neural network architecture to output prostate segmentations. PET and CT were co-registered and used as inputs to the model, with the 'manual' labels serving as ground truth segmentations. A weighted sum of boundary loss, Dice similarity coefficient (DSC), and binary cross entropy was used as the loss function of the model, while the predicted segmentations were evaluated using DSC and Hausdorff distance (HD).
Results: Our best prostate segmentation model was a multi-class segmentation neural network trained on both PET and CT modalities. The model achieved a mean HD of 7.21 ± 2.19, a mean DSC score of 0.82 ± 0.086, with a minimum of 0.52, a maximum of 0.90, and a median of 0.85 on the unseen test data from the three folds (n=30). The semi-automated 'threshold' segmentations achieved a mean HD of 9.06 ± 3.03 and a mean DSC score of 0.70 ± 0.18, with a minimum of 0.06, a maximum of 0.86, and a median of 0.74 across the manually labeled dataset (n=53). Our model outperformed the semi-automated segmentations in 24 of the 27 scans in the test set which had threshold labels available.
Conclusions: The proposed method is capable of accurately segmenting the prostate from a full-body PSMA PET-CT scan, with zero physician intervention. On average, this method outperforms existing semi-automated approaches that physicians might rely on. We believe this approach addresses a crucial step in the automation and subsequent standardization of prostate lesion detection and clinical reporting.