Abstract
4051
Introduction: <InvalidTag charset="UTF-8" />Radiopharmaceutical cocktails have been suggested and developed over the years to treat cancer. Cocktails of agents are attractive because one radiopharmaceutical is unlikely to have the desired therapeutic effect due to nonuniform uptake by the targeted cells (1). Therefore, multiple radiopharmaceuticals targeting different receptors of a cell is warranted (2). However, implementations in vivo have not met with convincing results due to the absence of optimization strategies. In this work, we present an optimizer, within the MIRDcell software platform, that can optimize a given cocktail of radiopharmaceuticals (drugs) in terms of the total activity needed to achieve a given surviving fraction (SF) of tumor cells. Using the methodology we present here, it is possible to determine the optimal drug combination for a sample of cells obtained from a patient and use that information to create a patient-specific cocktail to maximize the therapeutic effect for that patient using the least amount of total activity.
Methods: The optimizer package is developed as a feature for the MIRDcell software tool which is written in the Java programming language. The optimizer engine is based on the Sequential Least SQuares Programming (SLSQP) algorithm which is written in Fortran (3-5). We translated the collection of SLSQP subroutines from Fortran to Java. The Java SLSQP was integrated into our algorithm in MIRDcell that determines the molar activities that minimize the total activity of a cocktail of drugs needed to achieve a specified SF. This is done for each combination of drugs, subject to two constraints. The first constraint achieves the SF specified by the user. The second constraint requires that the total activity needed to achieve the SF with a given combination of drugs cannot exceed the activity required by any single drug comprising the combination. Validation of the optimizer package was done by reanalyzing our published data on suspensions of MDA-MB-231 human breast cancer cells treated with cocktails of four fluorochrome-labeled antibodies which were used to simulate cell killing with cocktails of 211At-labeled antibodies (drugs) (2).
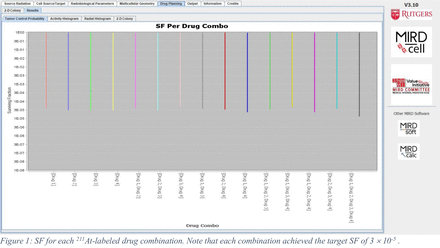
Results: <InvalidTag charset="UTF-8" />The optimized specific activities for each single 211At-labeled drug are in good agreement with the fixed values used in our publication (2) to cause the same SF. Not possible in our published analysis method (2), the MIRDcell algorithm determined the molar activities for each drug in each combination that are needed to achieve a specified SF (Figure 1) using the minimum total activity (Figure 2). Figure 2 shows that each combination achieved the required SF with different total activity and with different individual activities. The best combination was drug 1 + drug 2 + drug 3 with corresponding molar activities of 2.0x108, 1.3x107, and 9.5x106 GBq/mol. However, the simpler drug 1 + drug 2 combination with corresponding molar activities of 2.5x108 and 1.5x107 GBq/mol , respectively, was close behind.
Conclusions: The MIRDcell optimizer is capable of determining optimized drug combinations and corresponding molar activities needed to achieve a given SF. This approach could be used to analyze a sample of cells obtained from an animal or patient to predict the best combination of available drugs to be used in the treatment for maximum therapeutic effect with the least total activity.
Acknowledgement: This work was supported by NIH grant R01CA245139.
1. Neti et al. J Nucl Med. 2006;47:1049-1058.
2. Pasternack et al. J Nucl Med. 2014;55:2012-2019
3. Kraft D. A software package for sequential quadratic programming. DLR German Aerospace Center: Institute for Flight Mechanics; 1988. DFVLR-FB 88-28.
4. Kraft D. ACM Trans Math Softw. 1994;20:262–281.
5. Sequential Least SQuares Programming (SLSQP) algorithm. Version; 2019.