Abstract
3382
Introduction: The quantification of LV myocardial motion and wall thickening is widely regarded as crucial in the assessment of LV function. The objective of this work was to develop a 4D anatomically realistic display for the visualization of myocardial perfusion imaging (MPI)-derived wall thickening with CT templates of heart anatomy consistent with slice-wise thickening quantification.
Methods: Twenty-one Rb-82 PET rest/stress datasets were selected from our clinical databases of MPI studies and processed with the Emory Cardiac Toolbox (ECTb). Three anatomical templates of heart anatomy were selected from our database of clinical coronary CT angiography (CCTA) to represent small-, medium- and large-sized LV. For each CT template the LV endocardial and epicardial surfaces were subsequently modified to represent the end diastolic (ED) phase according to common assumptions on average LV wall thickness by imposing a constant myocardial thickness of 1 cm that tapers across the apical region to a minimum of 0.5 cm. For each gated PET study, the following data was extracted: the ED phase was identified, and ED LV dimensions extracted in terms of maximum longitudinal axis and max radius in mm; the 3D coordinates of LV mid-wall points were extracted for each time frame, hence delineating the LV wall 4D motion; for each point the thickening value (PETth) was computed; averaged values for each of the standard 17 segments were saved for validation. The 4D CT-derived surfaces were obtained as follows. The template was selected based on ED volume of each individual case and scaled to match the MPI-derived ED dimensions. From the 4D LV mid-wall points, a methodology based on a thin plate spline (TPS) technique was developed to analytically reconstruct the wall displacement fields from the ED to all other time frames. The resulting 4D motion was used to morph the CT ED template configuration into an analogous sequence of gated CT surfaces. Points thickening values were finally used to create epicardial and endocardial surfaces with a 4D motion equivalent to that extracted from the nuclear images. The point-by-point Euclidean distance (D) between each epicardial and endocardial CT surfaces was calculated and a mean value for each segment recorded. For each case, a geometrical thickening (GEOth) index was defined as (Dmax-Dmin)/Dmin over the cardiac cycle. To assess the potential of the dynamic CT surfaces to replicate the PET findings and results, subjects were separated in a normal and abnormal group and direct comparisons conducted between PETth and Geoth on a case-by-case basis, by individual segments and by pooling all segments. Pearson’s correlation coefficients (R) were calculated to assess the equivalence of the two measures.
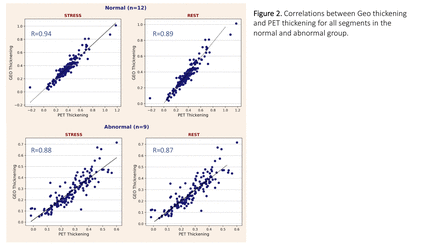
Results: Two subsets were created from the 21 selected subjects based on MPI SSS scores: a normal (SSS<4, n=12) and an abnormal group (SSS≥4, n=9). All datasets were processed and the procedure for the creation of the 4D CT display successfully applied to all cases. Calculated R coefficients were as follows: for all pooled segments Rstress and Rrest were respectively 0.94 and 0.89 (normal), and 0.88 and 0.87 (abnormal); when individual 17 segments were considered mean Rstress=0.92 [0.76-0.98] and mean Rrest=0.92 [0.79-0.98] for the abnormal group; mean Rstress=0.93 [0.75-0.98] and mean Rrest=0.90 [0.79-0.98] for the normal. When individual studies were considered, R was always >0.70 with the exception of 4 studies.
Conclusions: Our novel technique for the visualization of LV wall thickening by mean of 4D CT endocardial and epicardial surface models accurately replicated Rb-82 slice-wise wall thickening results showing promise of 4D display usage for diagnostic purposes.