Abstract
3345
Introduction: Clinical databases contain not only medical images but also accompanying free text reports. Information within these free text reports, such as clinical histories and physician interpretations, are generally not utilized in machine learning applications, often due to the reports’ unstructured nature as well as the uncertainty in how to best combine textual and image information. Here, we evaluate the ability of modern transformer-based natural language processing (NLP) methods to interpret text information in nuclear medicine clinical reports and explore multimodal learning as an approach to combine text and image information. We perform multimodal learning in the context of lymphoma 18F-fluorodeoxygluclose (FDG) PET/CT imaging and the prediction of visual Deauville scores (DS).
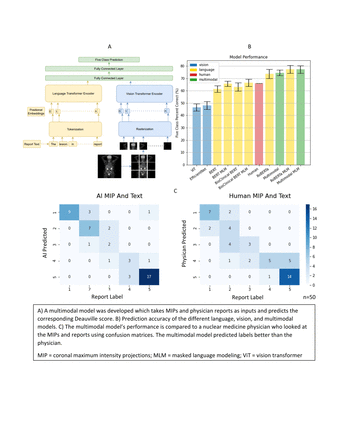
Methods: We extracted physician-assigned DS (ranging from 1 to 5: 1 is no uptake, 2 is uptake ≤ mediastinal blood pool, 3 is uptake ˃ mediastinal blood pool but ≤ normal liver uptake, 4 is uptake moderately more than normal liver uptake, and 5 is markedly increased uptake than normal liver uptake) from 1664 reports for baseline and follow-up FDG PET/CT exams. The DS were then redacted from the reports, and the remaining text was preprocessed with standard NLP cleaning techniques including synonym replacement, punctuation and date removal, and numerical rounding. The preprocessed reports were tokenized (i.e., split into subwords) and fed into one of three transformer-based language models: ROBERTA-Large, Bio ClinicalBERT, or BERT. To condition the models for nuclear medicine’s unique lexicon, models were pretrained with text reports using masked language modeling (MLM) in which 15% of the words in the reports were masked and the models would predict the missing word. The language feature vectors produced by the language models were then fed to a classifier and DS (1-5) were predicted. For vision, PET/CT images were converted into coronal maximum intensity projections (MIPs) with dimensions 384×384 and fed into a vision model, either ViT (a vision transformer) or EfficientNet B7 (a convolutional neural network). For the multimodal model, the outputs of the vision and language models were then concatenated and fed to a classifier. Monte Carlo cross validation (80% train, 10% validation, 10% test) was used for training and validation.To establish human level proficiency at this task as a benchmark for comparison, 50 exams were randomly selected and a nuclear medicine physician predicted DS based on coronal MIPs alone and then second based on the MIPs plus the radiology reports with DS redacted.
Results: We achieved 73.7% 5-class prediction accuracy using just the reports and the ROBERTA language model (linear weighted Cohen kappa κ=0.81), 48.1% accuracy using just the MIPs and the EfficientNet model (κ=0.53), and 74.5% accuracy using the multimodal model combining text and images (κ=0.82). When using MLM, ROBERTA improved from 73.7% to 77.4%, Bio ClincialBERT improved from 63.0% to 66.4%, BERT improved from 61.3% to 65.7%, and the multimodal model improved from 74.5% to 77.2%. The nuclear medicine physician correctly predicted the DS assigned in the clinical report just 58% of the time when using the MIP alone (κ=0.64), but improved to 66% accuracy when using the MIP and the report (κ=0.79).
Conclusions: We compared vision and language models in the context of classifying FDG PET/CT images according to visual DS. Pretraining the language models using MLM improved their ability to interpret clinical reports. We found marginal gains from combining language and vision models, but this is likely due to dominant prediction power of the language model relative to the comparatively weaker performance of the vision models. Overall, incorporating language into machine learning based image analysis is promising as modern language models are highly capable of interpreting language in the nuclear medicine domain.
Research support: This work was supported by GE Healthcare.