Abstract
3233
Introduction: Besides Alzheimer’s disease, Parkinson's disease (PD) is the most prevalent neurodegenerative disorder, affecting 2–3% of the old population over 65 years of age. Montreal Cognitive Assessment (MoCA) is a rapid nonmotor screening test that assesses different aspects of cognitive dysfunction. Early prediction of these symptoms may facilitate better temporal therapy, disease control, and identification of disease mechanisms. We set to investigate the prediction of MoCA in year 4 from year 0 & 1 imaging and non-imaging data, applying hybrid machine learning systems (HMLS), including features extraction algorithms (FEA) and feature selection algorithms (FSA) linked with regression algorithms (RA). Since our previous study focused on MoCA prediction via only clinical features (CF), this study plans to investigate the effect of radiomics features (RF) combined with CFs and conventional imaging features (CIF) to enhance prediction performance.
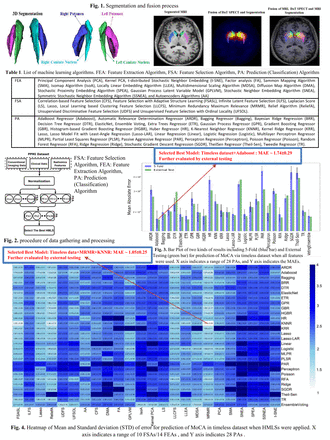
Methods: We selected 210 samples from the Parkinson's Progression Markers Initiative database. Further, 981 features, including CFs, CIFs, and RFs, were extracted from each dorsal striatum on DAT SPECT via the standardized SERA radiomics package. Four datasets (normalized by the z-score) were generated, namely using (i, ii) features in only year 0 (D1) or year 1 (D2), iii) longitudinal data (D3, putting cross-sectional datasets longitudinally next to each other), and iv) timeless data (D4, effectively doubling dataset size by listing both cross-sectional datasets separately). A range of optimal algorithms was pre-selected amongst various families of learner algorithms. First, we directly applied only RAs (28 different methods) on the datasets to predict MoCA in year 4. Subsequently, multiple HMLSs, including 14 FEAs or 10 FSAs followed by RAs, optimized by 5-fold cross-validation and grid search technique, were applied to the datasets to enhance prediction performances. 80% of all datapoints were applied to HMLSs to select the best model based on maximum performance resulting from 5-fold cross-validation. Subsequently, the remaining 20% was used for external testing of the selected model.
Results: When applying RAs without FSAs/FEAs to datasets, Adaboost achieved a minimum mean absolute error (MAE) of 1.74 ± 0.29 on dataset D4 in 5-fold cross-validation. Moreover, external testing performance of 1.71 confirmed our findings. Some algorithms, such as Least Absolute Shrinkage and Selection Operator (LASSO) and ElasticNet also had good Performances. Specifically, we achieved MAE of 1.93 ± 0.25 via D1 + ElasticNet, 1.77 ± 0.28 via D2 + LASSO, and 1.79±0.28, through D3+ElasticNet/ Histogram-based Gradient Boosting Regressor. When employing HMLSs (i.e. additional FEA or FSA step), Minimum Redundancy Maximum Relevance (MRMR) + K-Nearest Neighbor Regressor achieved the lowest MAE ~ 1.05 ± 0.25, selecting 92 relevant features (21 CFs +11 CIFs + 60 RFs) on dataset D4, significantly outperforming other HMLSs. Further, external testing performance of 0.57 also confirmed our finding. This is compared to our previous study (involving only clinical features), which achieved a minimum MAE of 1.68 ± 0.12 via non-dominated Sorting Genetic Algorithm+ Local Linear Model Tree algorithm. Overall, RFs added significant value to prediction of MoCA.
Conclusions: Our study shows the importance of using larger datasets (timeless), and utilizing optimized HMLSs, for significantly improved prediction of MoCA in PD patients. The best prediction performances were achieved by including RFs in the timeless dataset.