Abstract
2912
Introduction: Conventional spectrophotometric methods for protein estimation like Lowry’s and Bradford’s method are well documented and extensively used. An alternate method for protein estimation was standardized using the absorbance of the samples generated by HPLC. This could facilitate laboratories to double confirm the concentrations of the unknown specimens by two different methods. Conventional spectrophotometric methods for protein estimation like Lowry’s and Bradford’s method are well documented and extensively used. An alternate method for protein estimation was standardized using the absorbance of the samples generated by HPLC. This could facilitate laboratories to double confirm the concentrations of the unknown specimens by two different methods.
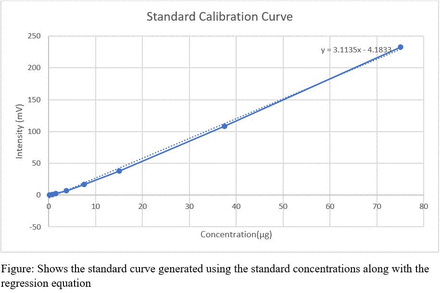
Methods: Standard solutions of the antibody (Trastuzumab) at varying known concentrations (0.15µg, 0.75µg, 1.5µg, 3.75, 7.5µg, 15µg, 37.5µg, 75µg) were prepared. HPLC was performed for each of these solutions and corresponding chromatograms obtained. The absorbance value for each of these concentrations was determined from the chromatogram. A calibration curve was then generated by plotting the concentration versus absorbance and the equation for best fit linear regression line generated. Blinded study was performed using three unknown concentrations and their absorbances determined by HPLC under identical conditions. The regression equation generated was used to determine the concentrations of these three unknown samples using their absorbance values obtained from the chromatogram.
Results: The absorbance values for the standard solutions as obtained by HPLC are listed in Table. Figure shows the standard curve generated using the standard concentrations along with the regression equation. An average % error of 5.3% was observed on comparing the original concentrations of the unknown samples with the calculated values.
The three unknown samples showed absorbances of 17.78mV, 105.01mV and 207.05mV corresponding to the concentration of 7.05µg, 35.07µg and 67.85µg respectively which closely matched with their respective concentrations (6.6µg, 33µg and 66µg). The concentrations of the unknowns were further validated using nanometer. The % error in measurement was 6.8%, 6.2% and 2.7% respectively.
Conclusions: In this study, it was feasible to generate a standard curve to estimate the protein concentration of unknown samples with good precision. This method of protein estimation could be an alternate or an added method to the conventional spectrophotometric methods.