Abstract
2421
Introduction: Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by progressive dysfunction of both upper and lower motor neurons in the brain and spinal cord. Previous research reported that GGGGCC hexanucleotide repeat expansion in the C9orf72 gene is the most frequent genetic cause of familial ALS. Some FDG-PET studies have focused on the causative gene in ALS patients with C9orf72, TBK1 mutation versus brain glucose metabolism. However, ALS genome-wide pathogenic gene mutations metabolic pattern has not been studied systematically. We aimed to replicate the metabolic changes previously described in a far larger group of patients with ALS and elucidate the difference of brain metabolic patterns between ALS patients with genetic and non-genetic in China mainland.
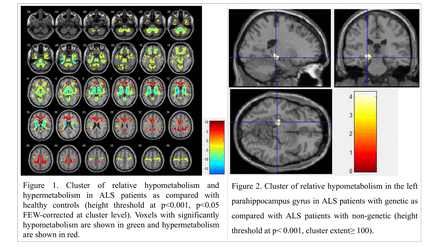
Methods: A total of 103 patients with ALS and 100 healthy controls underwent brain FDG-PET, and all ALS patients underwent detailed neurological examination and standardized electrodiagnostic examination. Sixty-five patients with ALS underwent genetic tests. C9orf72 and ATXN2 repeat expansions were detected by standard repeat-primed PCR. Moreover, other 49 ALS pathogenic genes also were screened by whole-exome sequencing (WES). Group comparison was carried out between ALS patients and healthy controls by using the two-sample t-test model of statistical parametric mapping (SPM 12), including the age at the time of PET imaging and sex as covariate. The height threshold was set at p<0.001, p<0.05 FWE-corrected at the cluster level. And we also performed a comparison between patients with and without genetic ALS, using the two-sample t-test model of SPM 12, including the age at the time of PET imaging, sex, duration, and ALS Functional Rating Scale-Revised (ALSFRS-R) as covariate. The height threshold was set at p < 0.001 uncorrected, cluster extent ≥ 100.
Results: Compared with healthy control participants, patients with ALS showed significant hypometabolism in the frontal lobe, temporal lobe, precentral gyrus, basal ganglia areas, midbrain and cerebellum, and hypermetabolism in the cingulate gyrus, frontal lobe, and occipital lobe. Of the patients who completed the genetic testing, fifteen patients carry pathogenic gene mutations and 50 patients with ALS don’t carry pathogenic gene mutations. Genetic ALS patients had lower Edinburgh Cognitive and Behavioural ALS Screen (ECAS) scores and relative hypometabolism in the left parahippocampus gyrus compared with those non-genetic ALS.
Conclusions: This large FDG-PET investigation provided strengthening evidence of relatively specific brain metabolic patterns in ALS patients as previously described. FDG-PET might also represent a potentially useful biomarker for ALS diagnosis in China mainland. Hypometabolism was found in the left parahippocampus gyrus in the genetic ALS patients as compared with the non-genetic ALS, suggesting a differential metabolic and cognitive-related state between the two conditions.