Abstract
1260
Objectives: We aimed to compare the results of different methods of spatial normalization of brain 18F-FDG PET data acquired on a hybrid PET-MRI system.
Methods: For the development of an institutional normal brain 18F-FDG PET-MR database we acquired additional brain PET and T1-weighted MR images in a clinical adult patient group who had whole-body 18-FDG PET examinations for pulmonary nodule evaluation. Exclusion criteria were history of neuropsychiatric disease, presence of visually apparent abnormalities and imaging artefacts on the acquired brain PET-MR images or any treatments that might confound brain metabolism. Brain PET-MR images were processed and analyzed using spm8. After manually correcting the image orientations in reference to the T1 MR template provided with spm8, T1 images were spatially normalized to the T1 MR template and the calculated deformation maps of patients were used for spatial normalization of the corresponding PET images. The spatially normalized images were smoothed using an isotropic 8 mm Gaussian filter. These data constituted the reference and used for the creation of an institutional 18F-FDG PET template. An additional patient group of mixed neurodegenerative disorders was selected to further characterize the effects of normalization template on quantitative results. All PET images were spatially normalized applying three PET templates (the 15O-water PET template provided with spm8, a neurodegenerative disease specific 18F-FDG PET template provided by Della Rosa P.A., et al. and the newly created institutional PET template) yielding three sets of PET data from the normal and neurodegenerative patient groups. Paired t-test was used to compare the three sets of normal group data with the reference normal database and independent samples t-test was used to compare the three sets of neurodegenerative group data with the reference normal database.
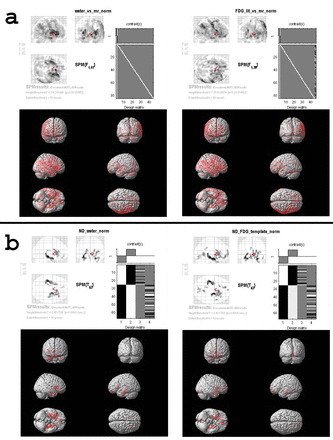
Results: Forty-two brain 18F-FDG PET-MR image sets were eligible for the creation of the institutional normal database and we had 25 patients in the neurodegenerative group. In the normal group no significant difference in metabolism was observed when institutional PET template was used for normalization (FWE-corrected p = 0.05 and extend threshold = 50). However, when water and neurodegenerative disease specific 18F-FDG PET templates were used for spatial normalization, significant differences were observed in extensive cortical regions and subcortical structures with a similar distribution (Figure 1a). In the neurodegenerative patient group, no significant metabolic differences in comparison to the normal database were detected when the institutional PET template was used for spatial normalization. However, the water PET template normalized data showed lower metabolism bilaterally in mesial temporal and orbitofrontal regions (Uncorrected p = 0.001 and extend threshold = 50). A similar result was observed when the neurodegenerative disease specific 18F-FDG PET template was used for normalization (Figure 1b).
Conclusion: Brain 18F-FDG PET images acquired on a hybrid PET-MR camera showed significant differences in metabolism when spatial normalization was done with different templates. The differences observed when PET data of the neurodegenerative patient group was compared with the normal database might be specific to hybrid PET-MR, since the distribution of hypometabolism corresponded to brain regions close to bony structures at the skull-base where the differences due to MR-based attenuation correction were expected to be maximum. Therefore, accurate quantitative analysis of brain 18F-FDG PET images acquired on a hybrid PET-MR system requires a separate PET template for spatial normalization and an institutional normal database for statistical analysis. Research Support: The PET-MR system used in this study was provided by Republic of Turkey Ministry of Development.