Abstract
2232
Introduction: Deep learning has been successfully applied on inter-frame subject motion correction for whole-body dynamic FDG PET to improve parametric imaging, with superior performance and faster computation than traditional methods. However, most current work considers only the image registration problem without taking tracer kinetics into consideration. We aim to incorporate Patlak regularization in an inter-frame motion correction deep learning network to directly optimize the Patlak fitting error and further improve model performance.
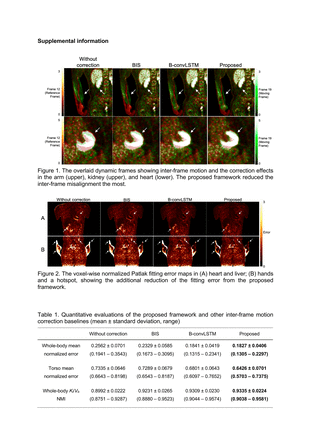
Methods: The dataset contains 27 subjects (22 cancer patients and 5 healthy controls). A 90-min whole-body dynamic continuous-bed-motion (CBM) FDG PET scan protocol on a Siemens Biograph mCT was used for each subject, including a single-bed chest scan in the first 6 minutes, the following 19 whole-body CBM frames, and individual input functions. 24 subjects are randomly selected for training and the remaining 3 are for testing. The proposed motion correction framework contains three modules: 1) the motion estimation module is a multiple-frame 3-D U-Net with a convolutional long short-term memory (LSTM) network layer combined at the bottleneck; 2) the spatial transformation module warps the input moving frame sequence with the estimated motion field; 3) the proposed analytical Patlak module that estimates Patlak fitting results and error with the motion-corrected frames and the individual input function. A Patlak loss regularization term using percentage fitting mean squared error (MSE) is introduced to the loss function in addition to an image similarity term measured by local normalized cross-correlation (NCC) and a displacement gradient loss penalizing local discontinuity. The network input is a sequence of 5-min frames (length = 5), each paired with the reference frame (Frame 12). An intensity cutoff with added Gaussian noise was implemented only for motion estimation to reduce the effects of high-intensity voxels and avoid local saturation. Following motion correction, the parametric images were generated by Patlak analysis with t* = 20 min at the original image resolution. We compared the motion correction performance with the traditional non-rigid registration baseline implemented in BioImage Suite (BIS) and a previously investigated deep learning network (B-convLSTM). The Patlak slope Ki and y-intercept Vb were overlaid to visualize motion-related mismatch. The normalized fitting errors in whole-body and torso as well as Ki/Vb normalized mutual information (NMI) were compared as the quantitative evaluation.
Results: The proposed framework further corrected the residual spatial mismatch in the dynamic frames and improved the alignment of Patlak Ki/Vb images. The voxel-wise normalized fitting errors are reduced in the Patlak error maps. Quantitatively, the proposed framework further reduced the normalized fitting errors and achieved higher Ki/Vb NMI. Both deep learning models substantially improved quantitative performance compared with BIS. Compared with B-convLSTM, the mean whole-body normalized fitting error further decreased from 0.1841 to 0.1827, the torso error additionally dropped from 0.6801 to 0.6426, and the whole-body mean Ki/Vb NMI increased from 0.9309 to 0.9335.
Conclusions: Incorporating Patlak regularization into the deep learning inter-frame motion correction framework has the potential to utilize FDG tracer dynamics and enhance network performance. The proposed framework has the potential to be extended as a joint end-to-end motion correction and parametric imaging model considering tracer kinetics.