Visual Abstract
Abstract
Attenuation correction remains a challenge in pelvic PET/MRI. In addition to the segmentation/model-based approaches, deep learning methods have shown promise in synthesizing accurate pelvic attenuation maps (μ-maps). However, these methods often misclassify air pockets in the digestive tract, potentially introducing bias in the reconstructed PET images. The aims of this work were to develop deep learning–based methods to automatically segment air pockets and generate pseudo-CT images from CAIPIRINHA-accelerated MR Dixon images. Methods: A convolutional neural network (CNN) was trained to segment air pockets using 3-dimensional CAIPIRINHA-accelerated MR Dixon datasets from 35 subjects and was evaluated against semiautomated segmentations. A separate CNN was trained to synthesize pseudo-CT μ-maps from the Dixon images. Its accuracy was evaluated by comparing the deep learning–, model-, and CT-based μ-maps using data from 30 of the subjects. Finally, the impact of different μ-maps and air pocket segmentation methods on the PET quantification was investigated. Results: Air pockets segmented using the CNN agreed well with semiautomated segmentations, with a mean Dice similarity coefficient of 0.75. The volumetric similarity score between 2 segmentations was 0.85 ± 0.14. The mean absolute relative changes with respect to the CT-based μ-maps were 2.6% and 5.1% in the whole pelvis for the deep learning–based and model-based μ-maps, respectively. The average relative change between PET images reconstructed with deep learning–based and CT-based μ-maps was 2.6%. Conclusion: We developed a deep learning–based method to automatically segment air pockets from CAIPIRINHA-accelerated Dixon images, with accuracy comparable to that of semiautomatic segmentations. The μ-maps synthesized using a deep learning–based method from CAIPIRINHA-accelerated Dixon images were more accurate than those generated with the model-based approach available on integrated PET/MRI scanners.
Accurately accounting for annihilation photon attenuation is essential for quantitative PET. In integrated PET/CT scanners, CT data are scaled to generate attenuation maps (μ-mapCT) that are used for PET attenuation correction. In integrated PET/MRI scanners, attenuation correction has been a challenge, as MRI does not directly provide information about tissue attenuation properties (1). The method initially implemented on one of the commercially available PET/MRI scanners, the Biograph mMR scanner (Siemens Healthineers), segmented the Dixon MR images into 4 compartments (i.e., background, lung, fat, and soft tissue) and assigned known linear attenuation coefficients to each of these classes to generate 4-compartment segmented μ-maps (μ-mapMR4C) (2). Because properly accounting for bone tissue attenuation is important, particularly in the pelvis, a model-based approach was subsequently developed to add bone tissue to the μ-mapMR4C. This whole-body 5-compartment model-based μ-map (μ-mapMR5C) generation approach uses a database of aligned MR images and bone segmentations for major body bones and involves coregistration of the subject’s MR image to the MRI model (3,4). The current method implemented on the Biograph mMR (software version VE11P) leverages the CAIPIRINHA (Controlled Aliasing in Parallel Imaging Results in Higher Acceleration)–accelerated Dixon 3-dimensional volumetric interpolated breath-hold examination sequence to acquire diagnostic-quality images with improved spatial resolution within the typical 18-s acquisition. In addition to providing diagnostic-quality images, this sequence was previously shown to improve the accuracy of the μ-mapMR4C (5).
Although the 5-compartment approach reduces the bias in the PET data quantification compared with the 4-compartment approach, it has several limitations when imaging the pelvis, the main focus in this work. First, air pockets (i.e., digestive tract gas) are difficult to identify and segment on the basis of the MRI data, leading to biased PET data quantification. Second, this attenuation correction method is prone to registration errors and does not account for the intra- and intersubject variability in bone density.
Deep learning–based methods are being rapidly adopted in the medical imaging field, with many applications in image segmentation (6–8), image registration (9,10), and image classification (11,12), among others. Such approaches that use convolutional neural networks (CNNs) and generative adversarial networks have also been implemented in PET and PET/MRI for various purposes, including synthesis of CT images for PET attenuation correction (or radiotherapy planning) (13–15). In the context of pelvic attenuation correction, deep learning–based methods have been applied to create synthetic CT images using Dixon MR and proton-density–weighted zero-echo time (16), standard Dixon (17,18), and T1-weighted LAVA Flex (GE Healthcare) water-only and T2-weighted MR images (19). All these studies reported improvements in the accuracy of μ-maps and reductions in bias in the reconstructed PET images, compared with those obtained using the standard segmentation-based μ-maps.
Previously proposed deep learning–based methods cannot synthesize accurate pseudo-CT images from pelvic MR images in the presence of air pockets, as they have an intensity similar to that of bone structures in the standard MR images. Furthermore, perfectly matched CT and MR images required for training of CNNs are not available since these images are acquired on separate scanners at different times. Therefore, the locations and sizes of the air pockets change between the 2 scans, leading to errors in both MRI–CT coregistration and image synthesis tasks. As an initial solution, Torrado-Carvajal et al. (17) filled the air pockets with values corresponding to soft tissue in the estimated μ-map images. Leynes et al. (16) filled the air pockets in the CT images with soft-tissue Hounsfield units (HUs) before training the CNN model. They reported artifacts in their pseudo-CT images due to assignment of bone HUs to air pockets. Both groups excluded the air pocket voxels from the PET data bias analyses. Alternatively, Bradshaw et al. (19) used a technique that involved an intensity-based threshold, morphologic closing, and manual adjustments to localize air pockets and place them on μ-maps.
In this work, we trained and evaluated CNNs to automatically segment air pockets from Dixon MR images and assessed the quantitative impact on the reconstructed PET images. Furthermore, we used the higher-quality CAIPIRINHA-accelerated Dixon images within a deep-learning framework to generate pelvic pseudo-CT maps, compared them with the μ-mapMR5C and μ-mapCT, and evaluated the impact of using these μ-maps on PET data quantification. Consideration of bowel gas positioning during the PET data acquisition will likely significantly impact accurate assessment of pelvic lesion uptake and have potential impactful clinical ramifications in both staging and longitudinal treatment assessment. Although outside the scope of the current study, our work therefore provides the technical foundation for future prospective studies to assess the impact of the proposed techniques.
MATERIALS AND METHODS
This retrospective study included data from 30 oncologic patients (age, 57 ± 10 y; 19 women and 11 men; weight, 69 ± 15 kg) who underwent successive, same-day PET/CT (as part of standard care) and PET/MRI (research) examinations. CAIPIRINHA-accelerated MRI Dixon data acquired from 5 additional subjects (age, 57 ± 5 y; 3 women and 2 men; weight, 72 ± 8 kg) were also included in this study and used only in the development and evaluation of the air pocket segmentation method. All patients gave written informed consent, and the local Institutional Review Board approved the study.
PET/MRI Data
Simultaneous PET/MRI data were acquired using the Biograph mMR scanner. Whole-body 18F-FDG PET data were acquired at 4 bed positions (injected dose, 568 ± 78 MBq) for 20 min approximately 2 h after radiotracer administration. Whole-body MRI data were acquired at 4 bed positions using the CAIPIRINHA-accelerated Dixon 3-dimensional volumetric interpolated breath-hold examination sequence (repetition time, 3.96 ms; first echo time, 1.23 ms; second echo time, 2.46 ms; flip angle, 9°; scan duration, 18 s) approximately 10 min after injection of a gadolinium-based MRI contrast agent (gadoterate meglumine [Dotarem; Guerbet]). This sequence provides in-phase and opposed-phase water and fat T1-weighted images that are typically used in the model-based μ-map estimation method. MR images were reconstructed with a voxel size of 2.1 × 2.6 × 2.1 mm.
CT Data
Low-dose CT data were acquired as part of the PET/CT acquisitions using Discovery 710 (GE Healthcare) (n = 26) and Biograph 64 (Siemens Healthineers) (n = 4) PET/CT scanners (voltage, 120 kV; tube current, 150 mA). The CT images were reconstructed with a voxel size of 0.98 × 0.98 × 5 mm. The CT data obtained from the 2 scanners were considered equivalent for the purpose of this study.
Image Processing
MR images were first corrected for low-frequency intensity nonuniformity using N4 bias correction (20). The scanner bed was removed from the whole-body CT images using intensity thresholding and morphologic operations. Subsequently, the pelvic region was manually cropped from the whole-body images. CT/MRI pairs were coregistered using affine and nonrigid transformations using NiftyReg (21). Finally, the images were resampled to generate a volume with 256 × 256 × n voxels. The voxel size of each volume was approximately 2.0 × 1.6 × 2.0 mm.
Air Pocket Segmentation
Air pockets present in the CT images were segmented using an image-thresholding algorithm (HUs < −700). Air pockets in the MR images were segmented using a region-growing algorithm implemented in ITK SNAP software (22). This semiautomatic procedure required manual placement of seeds on air pockets and editing of the resulting segmentations by experienced radiologists. These air pockets were used to train a CNN. A UNet (6,23) architecture with residual units, consisting of 4 down-sampling and 4 up-sampling layers, with rectified linear units used as the activation function, was chosen for this task. Three-dimensional MRI Dixon in-phase volumes were used as input data. The acquired images were resampled to an isotropic volume with a voxel size of 1 mm3, and multiple patches with a fixed matrix size of 96 × 96 × 96 voxels were extracted. To avoid overfitting during the training, data were augmented by applying ±10% image scaling and a random rotation with a ±10° angle. The MRI volumes were normalized to zero mean and unity variance. The network was trained and evaluated on a dataset of 35 subjects using a 5-fold cross-validation, where for each fold, the data were split into 80% training data (28 subjects) and 20% validation/testing data (7 subjects). The Dice similarity coefficient (DSC) was used as the loss function, and the network was trained using an Nvidia Tesla V100 graphics processing unit.
The accuracy of the segmentation network was evaluated by computing segmented air pocket volumes, DSC, and Hausdorff distance (24) at the 95th percentile between the segmentations obtained using the CNN and the semiautomatic methods. The DSC and Hausdorff distance are 2 metrics commonly used in evaluating image segmentation methods and are measures of similarity and largest segmentation error between 2 segmented regions. Volumetric similarity (25) was also computed using Equation 1:
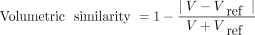 Eq. 1where V is the test volume and Vref is the volume of semiautomatically segmented air pockets.
Eq. 1where V is the test volume and Vref is the volume of semiautomatically segmented air pockets.
Pelvic Attenuation Map Synthesis
A separate network also based on the UNet (6) architecture was trained to synthesize pseudo-CT images from the 4 Dixon 2-dimensional axial images (17). Mean absolute error was used as the loss function. During the training, data were augmented by applying random displacements of 5 voxels and a random flip in the slices. The Dixon volumes were normalized to have zero mean and unity variance. A 5-fold cross-validation was performed where data were split to 80% training data (24 subjects) and 20% validation/testing data (6 subjects).
The HUs of the output pseudo-CT images were scaled to obtain the μ-maps (26). Voxels belonging to air pockets were assigned a linear attenuation coefficient of zero. μ-mapMR5C and μ-mapCT were also generated. All μ-maps were smoothed using a gaussian filter of 4 mm in full width at half maximum to match the resolution of the PET images. The percentage absolute and nonabsolute relative change (RC) were computed using Equation 2:
 Eq. 2where I is the test image and Iref is the reference image. CT-based μ-maps with CNN-derived air pockets were used as the reference image. Absolute and nonabsolute RCs were evaluated voxelwise in the whole pelvis and within 3 regions of interest (ROIs): bone, fat-based soft tissue, and water-based soft tissue. These ROIs were segmented using a thresholding algorithm on the ground truth μ-mapCT. Bones were obtained by excluding voxels with linear attenuation coefficients of less than 0.105 cm−1 and applying a flood-fill operation to capture the bone marrow. A water-based soft-tissue ROI was obtained by keeping only nonbone voxels within the 0.090–0.105 cm−1 range, and a fat-based soft-tissue ROI was obtained by selecting only voxels with linear attenuation coefficients in the 0.080–0.090 cm−1 range.
Eq. 2where I is the test image and Iref is the reference image. CT-based μ-maps with CNN-derived air pockets were used as the reference image. Absolute and nonabsolute RCs were evaluated voxelwise in the whole pelvis and within 3 regions of interest (ROIs): bone, fat-based soft tissue, and water-based soft tissue. These ROIs were segmented using a thresholding algorithm on the ground truth μ-mapCT. Bones were obtained by excluding voxels with linear attenuation coefficients of less than 0.105 cm−1 and applying a flood-fill operation to capture the bone marrow. A water-based soft-tissue ROI was obtained by keeping only nonbone voxels within the 0.090–0.105 cm−1 range, and a fat-based soft-tissue ROI was obtained by selecting only voxels with linear attenuation coefficients in the 0.080–0.090 cm−1 range.
Impact on PET Data Quantification
To evaluate the effects of using different μ-map generation methods on PET images, PET image reconstruction was performed using, first, a model-based μ-map with no added air pockets, as generated and used on the Biograph mMR scanner (μ-mapMR5C), and, second, MRI and CT-based μ-maps with CNN-predicted air pockets from MR Dixon images (μ-mapMR5C-CNNAIR, μ-mapMRDL-CNNAIR, and μ-mapCT-CNNAIR, respectively) (Fig. 1).
Overview of methodology. OSEM = ordered-subsets expectation maximization.
The PET images were reconstructed with the Siemens e7-tools (version VE11P) using the ordered-subsets expectation maximization algorithm (3 iterations and 21 subsets), with a voxel size of 2.1×2.1×2.0 mm, and smoothed using a postreconstruction gaussian filter with a full width at half maximum of 4 mm. Absolute and nonabsolute percentage RCs between the PET images attenuation-corrected using the μ-maps generated with the different methods were computed and reported for the whole pelvis and the ROIs listed above. To further study the effects of misclassified air pockets on the PET estimates in adjacent structures, a fourth ROI was obtained by dilating the semiautomatically segmented air pocket masks in all directions by 3 cm and subtracting the air pocket voxels from the dilated region.
RESULTS
Example Dixon in-phase MR images with air pockets semiautomatically segmented and predicted by the CNN algorithm are shown in Figure 2. The proposed method was able to segment both large- and small-volume air pockets and to distinguish between air pockets and other structures with low signal intensity on MR Dixon in-phase images, particularly bladder and bones, achieving a DSC of 0.75 ± 0.15, averaged across the testing/validation folds (Fig. 3A). Segmented air pocket volumes for each subject are shown in Figure 3B. The volumetric similarity between the 2 segmentations was 0.85 ± 0.14. Overall, there was no statistically significant difference between the air pocket volumes obtained using the 2 methods (paired t test, P = 0.30). Subject 30 had a significantly lower DSC and volumetric similarity than other subjects. This subject had one of the smallest volumes of air pockets, and the CNN misclassified the bladder as air, causing a large difference in segmented volumes (Supplemental Fig. 1; supplemental materials are available at http://jnm.snmjournals.org). The average 95th percentile Hausdorff distance between segmentations obtained with each method was 51.0 ± 52.4 mm.
Air pocket segmentation for representative subject (subject 21). Axial, coronal, and sagittal views of Dixon in-phase MRI are shown with semiautomatic segmentations of air pockets (red) and segmentation predicted by CNN (blue).
(A) DSCs between CNN-predicted and semiautomatic segmentations for 35 subjects. Horizontal line represents mean coefficient. (B) Volume of air in segmented regions obtained using semiautomatic and CNN approaches.
The μ-maps generated using model-based methods without (μ-mapMR5C) and with (μ-mapMR5C-CNNAIR) added air pockets, CT (μ-mapCT-CNNAIR), and the deep learning–based method (μ-mapMRDL-CNNAIR) are shown in Figure 4 for a representative subject. Qualitatively, the deep learning–based method appears to distinguish fat- and water-based soft tissue more accurately than the model-based method. Better representation of bone structures was also seen in μ-maps generated using the proposed method.
CT- and MRI-derived attenuation maps for representative subject (upper panels). CT-derived attenuation map with air pockets predicted by CNN was used as reference to compute corresponding RC maps shown in lower panels.
As shown in Table 1, the quantitative assessment confirmed these findings, μ-mapMRDL-CNNAIR being more similar to μ-mapCT-CNNAIR than was μ-mapMR5C-CNNAIR, with lower global and regional RCs. When all the voxels in the pelvis were compared, absolute RC was decreased from 5.1% to 2.6% when μ-maps were generated using the deep learning–based method rather than the 5-compartment model–based method. This difference was statistically significant (P < 0.001). The largest improvement was seen in the fat soft tissue, where the absolute RC was reduced by a factor of 2.6. The difference between absolute and nonabsolute RCs was statistically significant in fat-based soft-tissue ROIs and water-based soft-tissue ROIs. Although there were no significant group differences in the RCs obtained in the bones, the 5-compartment model-based approach failed to assign bone linear attenuation coefficients in most bones in 2 subjects (Supplemental Fig. 2).
Average Regional Nonabsolute and Absolute RCs Between Model-Based and Deep Learning–Based µ-Maps, Compared with CT-Based µ-Maps
PET images obtained using each μ-map and air pocket segmentation method and the corresponding RC maps with respect to PETCT-CNNAIR are shown for a representative subject in Figure 5. PETMRDL-CNNAIR had lower global RCs than did PETMR5C-CNNAIR. It can also be seen that PETMR5C had an area under the bladder with significantly increased 18F-FDG uptake, compared with the other PET reconstructions. This area corresponds to an air pocket misclassified as soft tissue in the μ-mapMR5C.
PET images reconstructed using CT- and MRI-based attenuation correction approaches (upper panels). RC maps for reconstructions performed using each method with respect to CT-based approach are shown in lower panels. Arrow indicates air pocket region that was incorrectly assigned to soft-tissue linear attenuation coefficients in μ-mapMR5C.
Averaged across all subjects, PETMR5C and PETMR5C-CNNAIR had larger nonabsolute and absolute RCs than did PETMRDL-CNNAIR, compared with PETCT-CNNAIR, both globally and regionally. Globally, the mean absolute RCs decreased from 7.1% to 4.9% and to 2.6% for PETMR5C, PETMR5C-CNNAIR, and PETDL-CNNAIR, respectively. As seen in Figure 6, improvements were also observed in all ROIs. PETMR5C had an RC of 20% and 12% in bone regions for the 2 subjects for whom model-based μ-maps did not include major bone structures.
Box plots of absolute (A) and nonabsolute (B) percentage change in reconstructed PET images using different attenuation correction and air pocket segmentation methods. Box plots were grouped for 5 ROIs. For each box, median is marked using central horizontal line and edges represent 25th and 75th percentiles of dataset. Whiskers were determined as 1.5 times interquartile range, and data points outside this range were identified as outliers. PETCT-CNNAIR was used as reference image in these calculations.
In the ROI surrounding air pockets, PETDL-CNNAIR had an absolute RC of 3.0% ± 1.4% (range, 0.1%–6.2%), whereas PETMR5C had an average absolute RC of 11.0% ± 6.5% (range, 0.7%–31.6%). In this ROI, PETMR5C images of 2 subjects had an absolute RC greater than 22%, as their μ-maps had large volumes of misclassified air pockets near the bladder.
DISCUSSION
Previous studies have highlighted the important role that PET (combined with both CT and MRI) plays in staging pelvic malignancies, planning chemoradiation, and assessing therapeutic response using multiple different tracers. With the recent Food and Drug Administration approval of PSMA-targeting agents, the clinical need for a reliable depiction of pelvic uptake using PET/MRI will only increase. In addition, PET/MRI is actively being explored for evaluation of inflammatory bowel disease (27). For all these applications, an accurate estimation of uptake will likely impact prognosis, choice of therapy, and treatment response assessment, therefore motivating our current study. First, misclassifying the air pockets as soft tissue could lead to false-positives due to overestimation of PET activity in these voxels. Second, lesions with increased uptake near air pockets could be missed because of the decreased lesion-to-background contrast. Third, the bias introduced in adjacently located lesions could impact the assessment of longitudinal changes. Finally, from a methodologic perspective, completely separating the air pocket segmentation from the pelvic attenuation map generation tasks when using deep learning approaches might increase the performance of the latter techniques because the related anatomic mismatches between the MR and CT images used for training could be eliminated (i.e., by filling the air pockets with soft tissue in both datasets).
The first aim of this work was to develop a deep learning–based approach to automatically segment air pockets in the pelvic region from high-resolution CAIPIRINHA-accelerated Dixon MR images. Semiautomatic segmentation of air pockets is a laborious and subjective process, especially when additional manual editing is required. The proposed CNN trained using semiautomatically segmented air pockets was able to accurately predict air pockets in new datasets with an average DSC of 0.75, suggesting it could be used to minimize this source of bias in the reconstructed PET images. Our results also showed that misclassifying air pockets as soft tissue can introduce bias in the reconstructed PET images, particularly in the adjacent structures, which could interfere with clinical interpretations (28).
High-resolution CAIPIRINHA-accelerated Dixon in-phase images were used in the delineation of air pockets to provide ground-truth data. However, these images contain a similar signal in air pockets and in some other structures such as bones, spinal cord, and some ligaments. Moreover, some of the subjects had a high number of small air pockets trapped between feces that were missed in the semiautomatic segmentation step but correctly identified by the CNN. Furthermore, CAIPIRINHA-accelerated Dixon in-phase images were acquired approximately 10 min after administration of gadolinium-based contrast agent, which caused the bladder to be separated into bright and dark areas, the latter being incorrectly classified as large air pockets in some subjects by the CNN. Acquisition of CAIPIRINHA-accelerated Dixon images before contrast agent administration will eliminate this issue and can potentially improve the performance of the air pocket segmentation method. Although these infrequent outliers could be corrected during the quality control step, the performance of the proposed method would likely increase if a larger number of datasets were available for training. In principle, this could be explored in future studies using only MR data. The segmentation method proposed could be combined with any pelvic μ-map generation approach to create maps that accurately reflect the physiologic state during the PET data acquisition.
Finally, although PET and MR images were acquired in a single scan using an integrated PET/MRI scanner, air pockets could have moved during the data acquisition because of peristalsis. In this study, we segmented the air pockets from a single CAIPIRINHA-accelerated Dixon acquisition and used the resulting μ-maps to attenuation-correct the PET data collected over a longer duration. One way to address this potential issue could be to repeat the CAIPIRINHA-accelerated Dixon acquisitions to detect potential air pocket movements over the course of the PET/MRI scan.
A second aim of this work was to train and test a separate CNN to generate more accurate pelvic μ-maps than those generated using the approach currently available on the Biograph mMR scanner (μ-mapMR5C). Qualitative and quantitative analyses indicate that a CNN trained with CAIPIRINHA-accelerated Dixon MR images is able to generate μ-maps with a better resemblance to μ-mapCT than μ-mapMR5C. We noticed that the overall absolute RC in the pelvis was reduced by a factor of 2, which was an improvement similar to that reported by Leynes et al. (16), Torrado-Carvajal et al. (17), and Pozaruk et al. (18). Compared with previous findings, we observed reduced differences in bony regions between the deep learning–based and model-based μ-maps. This reduction was due to the fact that the bone tissue is no longer misclassified as soft tissue in the μ-mapMR5C generated using the most recent method available on the Biograph mMR scanner.
The proposed image synthesis method uses a supervised CNN to perform a voxel-to-voxel regression of MRI intensities to CT HUs. This approach assumes perfect registration between the MR and CT images, which is hard to achieve. Our MRI and CT data were acquired on different scanners with differences in patient positioning, particularly in thigh flexion and rotation. Although we have used a combination of affine and nonrigid transformations to coregister the MRI and CT data of the training and validation datasets, some registration errors might still be present. Unsupervised learning techniques, such as the CycleGAN network incorporating cycle consistency loss function (29–31), can be used to alleviate the need for perfect alignment of MRI–CT pairs. However, these methods require access to larger pools of data for training, and they have to be properly validated for attenuation correction of PET data.
CONCLUSION
We developed a deep learning–based method to automatically segment air pockets from CAIPIRINHA-accelerated MR Dixon images. We also showed that a deep learning–based method can be used to synthesize μ-maps more similar to reference CT based μ-maps than the ones generated with the 5-compartment model-based approach as implemented commercially. Although our results suggest that this method might improve the CIs in studies requiring the use of quantitative PET metrics, additional studies on patients with pathologic changes are required to demonstrate its clinical utility.
DISCLOSURE
This work was supported in part by National Institutes of Health grant R01-CA218187. Ja Reaungamornrat and Ali Kamen are full-time employees of Siemens Healthineers. No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: Can we use CAIPIRINHA-accelerated Dixon MR images to automatically segment air pockets in the pelvic area and synthesize accurate pseudo-CT images for attenuation correction of PET data?
PERTINENT FINDINGS: A convolutional network to segment air pockets was trained and evaluated using CAIPIRINHA-accelerated Dixon images of 35 subjects. A separate network to synthesize pseudo-CT images was trained and tested using the Dixon images of 30 subjects who underwent sequential PET/CT and PET/MRI examinations. In a region surrounding the air pockets, an improvement by a factor of 3.7 was observed when PET images were reconstructed using deep learning–based μ-maps instead of standard model-based μ-maps.
IMPLICATIONS FOR PATIENT CARE: The proposed deep learning–based method can be used to accurately generate μ-maps with air pockets and can reduce the PET estimation bias in regions surrounding air pockets.
Footnotes
Published online July 22, 2021.
- © 2022 by the Society of Nuclear Medicine and Molecular Imaging.
REFERENCES
- Received for publication November 29, 2020.
- Revision received June 6, 2021.














