Abstract
Mesh-type and voxel-based computational phantoms comprise the current state of the art for internal dose assessment via Monte Carlo simulations but excel in different aspects, with mesh-type phantoms offering advantages over their voxel counterparts in terms of their flexibility and realistic representation of detailed patient- or subject-specific anatomy. We have developed PARaDIM (pronounced “paradigm”: Particle and Heavy Ion Transport Code System–Based Application for Radionuclide Dosimetry in Meshes), a freeware application for implementing tetrahedral mesh-type phantoms in absorbed dose calculations. It considers all medically relevant radionuclides, including α, β, γ, positron, and Auger/conversion electron emitters, and handles calculation of mean dose to individual regions, as well as 3-dimensional dose distributions for visualization and analysis in a variety of medical imaging software. This work describes the development of PARaDIM, documents the measures taken to test and validate its performance, and presents examples of its uses. Methods: Human, small-animal, and cell-level dose calculations were performed with PARaDIM and the results compared with those of widely accepted dosimetry programs and literature data. Several tetrahedral phantoms were developed or adapted using computer-aided modeling techniques for these comparisons. Results: For human dose calculations, agreement of PARaDIM with OLINDA 2.0 was good—within 10%–20% for most organs—despite geometric differences among the phantoms tested. Agreement with MIRDcell for cell-level S value calculations was within 5% in most cases. Conclusion: PARaDIM extends the use of Monte Carlo dose calculations to the broader community in nuclear medicine by providing a user-friendly graphical user interface for calculation setup and execution. PARaDIM leverages the enhanced anatomic realism provided by advanced computational reference phantoms or bespoke image-derived phantoms to enable improved assessments of radiation doses in a variety of radiopharmaceutical use cases, research, and preclinical development. PARaDIM can be downloaded freely at www.paradim-dose.org.
Radiopharmaceutical use demands consideration of dosimetric implications in clinical use cases, preclinical animal studies, and cell-based assays. For human use, accurate dosimetry is a requisite for meaningful risk projection from low-dose imaging procedures, as well as for reliable pre–radionuclide therapy dose planning, response assessment, and characterization of potential toxicities. In the preclinical setting, the considerations are similar but more nuanced. For example, the high activities per unit mass used in preclinical PET and SPECT investigations generally deliver elevated absorbed doses to small-animal hosts and thus have the potential to induce radiotoxicity, alter gene expression, or present other biologic anomalies complicating interpretation of the scientific output. Tools have been developed to perform organ-level dosimetry in humans and mice (1) but generally implement a limited set of phantoms and have limited or no provisions for assessment of nonuniform dose deposition within phantom regions. Alternatively, several image-based dosimetry tools exist (2–5), but their accuracy is dependent on the quantitative accuracy of PET/SPECT imaging. A versatile methodology that combines the advantages of both approaches would be beneficial. Finally, the dose to cell components has been shown to be sensitive to the cell size or shape and to the location of the decay site (6,7), particularly for radiation with high linear energy transfer. MIRDcell (8) has been a reliable tool for cell-level dose assessment for some time and models the dose to single cells or cell clusters. However, all cells within MIRDcell are modeled as concentric spheres, which in certain cases can be limiting. Considering the increasing emphasis on high-linear-energy-transfer radionuclide therapies, a practical option for cell-level dose modeling to irregular cell geometries would be timely.
We have developed PARaDIM (pronounced “paradigm”: Particle and Heavy Ion Transport Code System [PHITS]–Based Application for Radionuclide Dosimetry in Meshes), a program that uses PHITS version 3.07 or later (9,10) for dose computation. No knowledge of the PHITS source code, input file format, or parameter syntax is required for its use. Similar to other programs for mean organ-level dose assessment based on precomputed S values, PARaDIM implements a user-friendly graphical user interface for input of time-integrated activity coefficients (11) and generates organ-level mean absorbed doses under the assumption that the activity is uniformly distributed throughout each source region. Additionally, PARaDIM provides voxelized 3-dimensional (3D) dose maps at a user-specified resolution, supporting assessment of heterogeneity of dose deposition. PARaDIM supports cell-level dosimetry based on modeling single cells or cell clusters of arbitrary shape or spacing and with arbitrary components through characterization of cells and clusters as a tetrahedral mesh.
It is our expectation that PARaDIM will allow researchers in preclinical nuclear medicine to make improved assessments of radiation dose in small-animal imaging and therapy studies and cell-based assays, as well as provide more accurate projections of absorbed dose to potential patients of various body habitus in support of radiopharmaceutical development and investigational new drug applications. For human dosimetry, PARaDIM will provide a robust supplement to image-based methods, primarily offering a level of nonuniform dose assessment applicable to patient populations and personalized output from patient-specific phantoms derived from tomographic image data.
MATERIALS AND METHODS
Code Development and Functionality
PARaDIM is provided as a stand-alone tool executable for Microsoft Windows operating systems. The source code was entirely written in Python (version 3.6; www.python.org) within the PyCharm integrated-development environment (https://www.jetbrains.com/pycharm/). The graphical user interface was created using Python’s built-in Tkinter and Ttk libraries, and the external libraries Numpy (12) and NiBabel (13) are utilized for select data-processing functions. The code was compiled using PyInstaller (www.pyinstaller.org). Each portion of the code was tested with PHITS versions 3.07–3.10+ on Windows 7 and Windows 10 platforms, as the code was developed.
PHITS is a multipurpose particle-transport code handling transport of virtually all known particle types for applications in medical physics and radiation protection, among other fields. PARaDIM assembles user-specified phantom geometry, source definition, and simulation parameters into a PHITS-compatible input file for execution (Fig. 1). After submission of a simulation from PARaDIM, PHITS initiates and transports the various particle and photon emissions within the phantom geometry and tallies energy deposition within phantom regions or in a voxel grid.
Flowchart describing function of PARaDIM for organ-level and 3D dose calculation and relationships with relevant techniques. CAD = computer-aided design; MIP = maximum-intensity projection.
For all simulations, PARaDIM uses tetrahedral mesh-type computational phantoms (14–19). Yeom et al. (18) provide a detailed discussion of computation speeds and memory requirements of this phantom format relative to voxel phantoms. The PHITS code was recently updated to support such phantoms, and their use within PHITS for absorbed dose calculations has been well validated (14). A library of tetrahedral phantoms has been provided with PARaDIM (Table 1). Phantoms can be accessed through PARaDIM’s “Phantom Selection” module (Fig. 2). Importantly, user-defined phantoms are also supported, including, for example, personalized phantoms derived from segmentations of data from tomographic anatomic imaging modalities (e.g., CT or MR) that have been converted to the standard tetrahedral *.ele/*.node format.
Preconfigured Tetrahedral Phantoms Available for PARaDIM
Snapshot of “Phantom Selection” module in PARaDIM. Here user may select phantom from library or may add phantom to standard library via “Import New Phantom” function. Tissues and masses for currently selected phantom may be viewed or exported. Phantoms may be scaled to particular mass by providing entry in “Scale to Mass” input field.
The calculation type, radionuclide, and simulation options (e.g., resolution and physical model options) are specified in the “Parameter Setting” module. Three general types of calculation can be configured: absorbed dose, dose rate, and computation of S values to be used in outside calculations of mean region-level absorbed dose. Each output mode may be calculated at the tetrahedral region level or in 3 dimensions (voxel level). The EGS5 mode (20) of PHITS is used for positron, negatron/electron, and photon transport; the default PHITS algorithm is used for α-decay when applicable. The accuracy of the EGS5 mode of PHITS in calculating dose-point kernels for monoenergetic electrons and β-emitting isotopes has been well validated (21). PARaDIM does not automatically account for decay of radionuclide progeny; that is, the user must therefore ensure that if the radionuclide is part of a decay chain, the time-integrated activity coefficients for the progeny are characterized appropriately and simulated separately. Energy deposition arising from recoiling daughter nuclei is not scored.
Source particles for each phantom organ are generated using the PHITS multisource method. Particle type is sampled on the basis of weighted intensities defined by the PHITS radioisotope source libraries (equivalent to International Commission on Radiological Protection [ICRP] publication 107) (22), and the particle is given an initial energy sampled from the same libraries (full spectrum for β-particles and monoenergetic spectrum for photons/Auger electrons/internal conversion electrons/α-particles). For radionuclides with multiple emission types, the user may specify which particles to include in the simulation. Statistical uncertainties η for voxels or tetrahedral regions are calculated using the “History Variance Mode” of PHITS, where the relative error is given as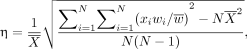 Eq. 1
Eq. 1
N is the total history number; 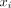 and
and  are tally results and the source weight of each sample, respectively; and
are tally results and the source weight of each sample, respectively; and  and
and  are the respective mean values of the tally results and source weights of N samples.
are the respective mean values of the tally results and source weights of N samples.
For the default absorbed dose calculation mode of PARaDIM, the source intensities are defined by whole-region time-integrated activity coefficients specified by the user within the “Source Definition” module. For the dose rate calculation mode, input is given as either activities in source regions or fraction of administered activity in source regions. To configure an S value calculation, one simply specifies the source region.
Several import–export and data-processing options are provided to streamline workflow and, particularly for 3D mode, enable visualization and analysis (e.g., dose–volume histogram generation) in numerous types of imaging software. Organ-level absorbed doses that are output from PHITS are viewable in the PARaDIM graphical user interface for convenient readout.
Example Dose Calculations in Human Phantoms
Dosimetric estimates for an 18F-labeled reference PET tracer, 18F-AlF-peptide, were obtained separately in PARaDIM and OLINDA 2.0 for the reference adult Korean man and woman using time-integrated activity coefficients extrapolated from mouse biodistribution data (Table 2 and supplemental information [supplemental materials are available at http://jnm.snmjournals.org]). For these comparisons, male and female Mesh-Type Reference Korean Phantoms (MRKPs) (17) were used in PARaDIM, whereas the adult male and female ICRP 89 (23) and legacy Oak Ridge National Laboratory phantoms (24) were used in OLINDA with organ weights scaled to equivalent masses of the MRKPs. For organs in the MRKPs that are subdivided (e.g., kidney, comprising separate tetrahedral regions defining the renal cortex, medulla, and pelvis as opposed to the entire kidney), the time-integrated activity coefficient was allocated to each constituent component as the product of the component’s mass fraction and the total time-integrated activity coefficient for the whole organ. One million total source particles were simulated in each phantom, and PARaDIM defaults were used for all other parameters.
Time-Integrated Activity Coefficients Used for Absorbed Dose Calculations in PARaDIM and OLINDA 2.0, Calculated for Male and Female MRKP Based on Mouse Biodistribution of Reference Peptide
Organ-level mean absorbed doses for whole organs uniquely defined in the MRKPs (e.g., liver and brain) were output directly from PARaDIM. To facilitate comparison of the results of PARaDIM with those of OLINDA 2.0, whole-organ mean absorbed doses  for target organs that are subdivided into components
for target organs that are subdivided into components  of mass
of mass 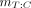 were calculated as
were calculated as Eq. 2
Eq. 2
For further comparison and validation independent of the distribution of decays and geometry, the supplemental information provides S values for selected sources and targets for 18F in the MRKPs, which enables comparison of PARaDIM with other Monte Carlo transport codes.
Additionally, dose rates were calculated with PARaDIM in a phantom derived from segmentation of a PET/CT dataset (data obtained from the Cancer Imaging Archive anti–PD-1 melanoma collection) (25), demonstrating the use of patient-specific phantoms in PARaDIM. The parameters for this simulation were as described above.
Example Dose Calculation in Small-Animal Phantoms
The mouse whole-body (MOBY) phantom has been implemented by several investigators for mouse dosimetry calculations (26–28). Using methodology described in the supplemental information, MOBY was converted to tetrahedral geometry to enable its use within PARaDIM; 18F self–S values for MOBY’s constituent organs were calculated and compared with previous literature that used voxel-based methods with this phantom (27). One million source particles were simulated separately from each organ, resulting in less than 1% relative uncertainty for each self–S value. Additionally, decay of 18F, 90Y, and 186Re was simulated from MOBY liver sources in 3D mode (106 events, 0.75-mm resolution).
Example Cell-Level Dose Calculations
PARaDIM was compared with MIRDcell for validation of accuracy of absorbed dose computation at the cell level. MIRDcell models cells as concentric spheres, with the radii of the cell nucleus and the complete cell specified in integer increments of micrometers. The cell membrane, cytoplasm, nucleus, or entire cell may be used as sources or targets. Selected S values for cells of various dimensions were generated with MIRDcell and compared with those computed with PARaDIM using a series of tetrahedral cell phantoms constructed with equivalent dimensions (supplemental information). One million source particles were simulated for 18F and 211At using PARaDIM’s parameter defaults; calculations with MIRDcell used the full β-spectrum when applicable.
In contrast to MIRDcell, PARaDIM is not limited to cells modeled as concentric spheres with regular spacing. Within the constraints of computation time and memory, detailed arrays of cells with complicated geometry, arbitrary size, and nonuniform spacing may be defined as a tetrahedral mesh and used in dose calculations. As an illustration, a calculation for a hypothetical cell geometry similar to one encountered commonly in in vitro cell radioassays was designed. A pellet of 500 nonspheroidal cells contained in a 1.5-mL centrifuge vial in 370 μL of medium was modeled as a tetrahedral mesh; 37 kBq of total 64Cu activity was assumed to be distributed as follows: 9.5% within the cytoplasm of the cells, 0.5% within the cell nuclei, and 90% in the surrounding medium.
RESULTS AND DISCUSSION
PARaDIM is a flexible tool for dosimetry in many settings relevant to nuclear medicine and radiation protection, yielding estimates of region-level and nonuniform absorbed doses using a tetrahedral mesh phantom to model the relevant anatomy or geometry. Whereas many software programs implement only one or a limited set of phantoms, a motivating factor for using PARaDIM is the ability to easily simulate dose in novel phantoms. It is designed to integrate well with modern imaging software for output postprocessing and visualization and with workflows for development of bespoke phantoms—for example, from segmentation of medical imaging data (Fig. 3). Figure 4 gives a cell-based example that is necessarily situational but further illustrates PARaDIM’s utility for dose calculation in truly unique or customized settings, especially when allied with computer-aided design workflows for custom phantom generation (this is facilitated by PARaDIM’s use of tetrahedral mesh phantoms). Other use-cases in which PARaDIM excels include advanced phantoms for which complex source and target regions are detailed, computational reference phantoms for which S values have not yet been calculated, and reference phantoms that have been deformed (e.g., to match a particular subject’s geometry). PARaDIM is also capable of dose calculation in brachytherapy and certain external-radionuclide-exposure scenarios. Such calculations can be accomplished, for example, by defining brachytherapy or external sources along with the biologic system together within a single tetrahedral mesh (Supplemental Fig. 2); however, validation of the program for these purposes is outside the scope of the present article. Finally, 4-dimensional dose calculations may be conducted in PARaDIM by importing a dynamic geometry as separate static frames.
Conceptual example of patient-specific, tomographic imaging–derived phantom use in PARaDIM; quantification of dose rate at scan time during restaging with 18F-FDG. (A) 18F-FDG PET maximum-intensity projection and volume-rendered CT. (B) Slice and 3D-rendered view of CT-based segmentation. (C) Quantification of 18F activity in coregistered PET volume within each segment. (D) Maximum-intensity projection and slice view of 3D output following dose rate simulation in PARaDIM (1 × 106 events simulated). (E) Mean organ-level dose rate coefficient output from PARaDIM.
Cell-level dose rate calculation for step of in vitro radioassay. (A) 3D rendering of system. (B and C) Dose rate scored at voxel level for cell array (6-μm isotropic voxel size, 1 × 107 total history number): maximum-intensity projection (B) and slice view with cell boundary overlay (C).
Agreement of PARaDIM with Other Methods
We have compared absorbed doses computed with PARaDIM to 2 widely used and well-validated applications for internal dosimetry: OLINDA 2.0 and MIRDcell. Although the validation presented is not exhaustive, the agreement provides some confidence in PARaDIM’s results and illustrates the value of PARaDIM as both a stand-alone dosimetry tool and a complement to these well-established methods. Human organ-level absorbed doses calculated with PARaDIM and OLINDA agreed well, within approximately 20% for most organs, with differences attributed primarily to geometric and spatial differences between the MRKPs used in PARaDIM and the ICRP 89 series and stylized Oak Ridge National Laboratory reference human phantoms used in the latter program (Fig. 5). The most significant differences observed were for organs comprising separate wall and content components (e.g., urinary bladder wall and heart wall), for which doses computed with OLINDA were higher than those computed with PARaDIM. These differences were not unexpected, as OLINDA calculates doses to the wall target regions  from content sources
from content sources  by assuming that the specific absorbed fraction
by assuming that the specific absorbed fraction 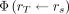 for β-radiation is approximated by the following (30):
for β-radiation is approximated by the following (30): Eq. 3which is known to yield conservatively high estimates for low-energy β-emitters, including 18F. The corresponding 3D output of PARaDIM for these simulations is shown in Figure 6.
Eq. 3which is known to yield conservatively high estimates for low-energy β-emitters, including 18F. The corresponding 3D output of PARaDIM for these simulations is shown in Figure 6.
Absorbed dose estimation for intravenous administration of 18F-AlF-peptide in reference Koreans: comparison of organ-level absorbed dose coefficients computed with PARaDIM vs. OLINDA 2.0 for males (top) and females (bottom). Asterisks represent tissues not defined in respective phantom. AF = adult female; AM = adult male; LLI = lower large intestine; ULI = upper large intestine; ORNL = Oak Ridge National Laboratory; SI = small intestine; UB = urinary bladder.
Absorbed dose coefficients and corresponding relative uncertainties scored at voxel level in MRKPs, with phantom geometry juxtaposed for anatomic reference. AF = adult female; AM = adult male; MIP = maximum-intensity projection.
Self–S values for the MOBY phantom showed good agreement with previous work (Table 3) (26); overall agreement was within 10% for all organs, but the S values computed with PARaDIM showed a positive bias of approximately 4% on average, expected to be due in part to differences in algorithms (GEANT4 vs. PHITS-EGS5) and to geometric and partial-volume considerations arising from the use of voxel versus tetrahedral mesh methods.
The agreement with MIRDcell for cellular S value calculations was similarly good (<10%) for both high-linear-energy-transfer emitters and low-linear-energy-transfer emitters, with the largest differences arising for the nucleus self–S value (i.e., S(N←N)) (Supplemental Table 3). The reason for this difference is expected to be primarily the difference in the algorithms and cross-sectional data used, with MIRDcell using an analytic method based on stopping power and geometric factors (31), in contrast to PARaDIM’s use of PHITS.
S Values for 18F in MOBY Phantom in Comparison to Those Reported by Keenan et al. (26)
Computational Speed
Computation times in PARaDIM are variable, influenced heavily by the particle history number, the level of detail of the phantom, the spatial dimensions of the phantom, the selected output resolution, the decay modes and energies of the radionuclide simulated, the energy cutoffs, and the computer hardware and architecture used. Generally, PARaDIM does not require supercomputing clusters or other expensive computational assets for most simulations. By default, calculations are run on a single core, but both shared-memory parallel processing and manual parallel processing are supported for acceleration. For reference, the MRKP simulations described here required 3–4 h of computation time for a single-core calculation on a standard desktop workstation (3.6-GHz Intel Xeon 5122 processor, Windows 10 operating system). In contrast, the computation time for the cell S value calculations given the same particle history number (106) was generally less than 5 min, because of the reduced size and complexity of the cell phantoms used.
Advantages and Disadvantages of the Multisource Technique Relative to the S Value Approach
Although dosimetry calculations based on precomputed S values produce essentially instantaneous results, in any novel phantom the S values must first be determined. Relative to the en masse multisource technique that PARaDIM is capable of using, calculation of S values for each individual tissue in a phantom, to the same degree of relative uncertainty, is computationally intensive and requires a vast amount of oversampling if the source distribution is unknown. The reason is that, for most tissues, the absorbed dose contribution from decays occurring in distant sources or sources with a low density of decays is generally dominated by that from 1 or 2 hot sources or that imparted from decays occurring within or adjacent to the target. Furthermore, in the en masse method of calculation, sampling statistics are necessarily increased in regions with elevated dose, where accuracy is often more important; in this respect, however, attention must be paid to the individual tissue radiation weighting factors. PARaDIM does not incorporate such radiation weighting factors (i.e., only absorbed dose is scored, not dose equivalent), but if needed, weighting factors may be applied to the output in other standard software (e.g., Microsoft Excel).
Evaluation of Nonuniform Dose Deposition with PARaDIM
Nonuniform dose deposition arising from sources with homogeneous distributions of radioactivity may be evaluated in 3D mode within structures that can be sufficiently resolved at the specified voxel size and for radiation with a penetration range that is long in comparison to the voxel size. As an example, absorbed dose from MOBY phantom liver sources was calculated in 3D mode (with 0.75-mm isotropic voxel dimensions) using 3 radionuclides with largely different mean β-emission energies (Fig. 7). The mean soft-tissue range of β-radiations emitted from 18F and 186Re are only 0.6 and 1.0 mm, respectively. Because these ranges are not long in comparison to the voxel size, in this case this resolution is largely unsuitable for evaluating variation in deposited dose from β-emissions from either 18F or 186Re due to partial-volume effects. Dose–volume histograms were derived from the 3D dose distributions to help illustrate this point; differences in the respective normalized dose–volume histograms for 18F and 186Re (Fig. 7C) are not meaningfully different, despite the large relative difference in mean β-energy and range between these radionuclides. Conversely, the mean range of β-emissions from 90Y is 3.9 mm, which is large relative to the voxel dimensions, and the variation in dose within the liver and dose to organs surrounding the liver could be meaningfully assessed by defining regions or volumes of interest or, globally, within organs using a dose–volume histogram. Similarly, 511-keV photons produced by 18F positron annihilation are of very long range in this respect, and the variation in dose deposition at sites distant from the liver is evident (Fig. 7A); this effect is absent from the 186Re and 90Y simulations because of their low abundance of energetic photons.
Evaluation of inhomogeneous dose deposition in MOBY phantom with PARaDIM. (A) Radionuclides with increasingly energetic β-emissions simulated from liver sources. Note shallow gradient of dose deposition at liver boundaries (magenta outline) and significant dose to adjacent organs with 90Y, in comparison to shorter-range β-emitters 18F and 186Re. (B) Dose–volume histograms for liver, for each radionuclide. (C) Dose–volume histograms from B normalized to maximum-dose voxel. SI = small intestine.
Alternatively, nonuniform doses within phantom regions may be evaluated to an increased extent by dividing regions into subregions using mesh editing software; the mean dose within each subregion would then be tallied individually in PARaDIM. This approach avoids the partial-volume effects associated with voxel-format dose tallies.
CONCLUSION
Advancements in medical image segmentation, 3D geometric modeling/manipulation/representation/visualization, and Monte Carlo particle transport simulation have enabled accurate calculation of absorbed doses in tetrahedral mesh phantoms that intricately characterize the anatomy of subjects, including humans, animals, and cells. The use of such phantoms in dosimetric simulations has previously been restricted largely to investigators with an advanced understanding of select Monte Carlo transport codes. PARaDIM extends the benefits of this powerful simulation technique to all investigators in the field, without the need for advanced computer expertise, and can be obtained freely at www.paradim-dose.org, along with its terms of use, disclaimer, and documentation.
DISCLOSURE
This work was supported by the Memorial Sloan Kettering Radiochemistry and Molecular Imaging Probes Core, which was supported in part by National Institutes of Health grant P30 CA08748. This work was also supported by NIH grant R35 CA232130, the Mr. William H. and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and by the Center for Experimental Therapeutics of Memorial Sloan Kettering Cancer Center (Jason Lewis). Lukas Carter acknowledges support from a Ruth L. Kirschstein NRSA postdoctoral fellowship (NIH F32 EB025050). No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: Can we facilitate the use of advanced methods in Monte Carlo–based internal dosimetry by scientists and clinicians?
PERTINENT FINDINGS: PARaDIM, a user-friendly graphical user interface for internal dose calculations via PHITS, was developed, tested, and validated in this work.
IMPLICATIONS FOR PATIENT CARE: PARaDIM will benefit the field by facilitating setup of internal dose calculations with modern computational reference phantoms and subject-specific phantoms derived from anatomic imaging, offering an advanced alternative to existing methods for radiation dose assessment.
Footnotes
Published online Jun. 14, 2019.
- © 2019 by the Society of Nuclear Medicine and Molecular Imaging.
REFERENCES
- Received for publication March 22, 2019.
- Accepted for publication May 29, 2019.














