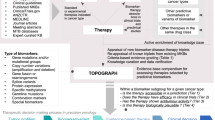
Abstract
This Perspective highlights biomarkers that are expressed as a consequence of cancer development and progression. We focus on those biomarkers that are most relevant for identifying patients who are likely to respond to a given therapy, as well as those biomarkers that are most effective for measuring patient response to therapy. These two measures are necessary for selecting the right drug for the right patient, regardless of whether the setting is in drug development or in the post-approval use of the drug for patients with cancer. We also discuss the innovative designs of clinical trials and methodologies that are used to validate and qualify biomarkers for use in specific contexts. Furthermore, we look ahead to the promises and challenges in the field of cancer biomarkers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Luo, J., Solimini, N. L. & Elledge, S. J. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136, 823–837 (2009).
Polyak, K. & Weinberg, R. A. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nature Rev. Cancer 9, 265–273 (2009).
Paul, S. M. et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nature Rev. Drug Discov. 9, 203–214 (2010).
Polyak, K., Shipitsin, M., Campbell-Marrotta, L., Bloushtain-Qimron, N. & Park, S. Y. Breast tumor heterogeneity: causes and consequences. Breast Cancer Res. 11 (Suppl. 1), 18 (2009).
Polyak, K. & Kalluri, R. The role of the microenvironment in mammary gland development and cancer. Cold Spring Harb. Perspect. Biol. 2, a003244 (2010).
Pegram, M. D., Pauletti, G. & Slamon, D. J. HER-2/neu as a predictive marker of response to breast cancer therapy. Breast Cancer Res. Treat 52, 65–77 (1998).
Druker, B. J. Perspectives on the development of imatinib and the future of cancer research. Nature Med. 15, 1149–1152 (2009).
Vultur, A., Villanueva, J. & Herlyn, M. Targeting BRAF in advanced melanoma: a first step toward manageable disease. Clin. Cancer Res. 17, 1658–1663 (2011).
Chapman, P. B. et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 364, 2507–2516 (2011).
US Food and Drug Administration. FDA labeling information — Zelboraf. FDA website [online], (2011).
Soda, M. et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566 (2007).
Kwak, E. L. et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 363, 1693–1703 (2010).
Cui, J. J. et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J. Med. Chem. 54, 6342–6363 (2011).
US Food and Drug Administration. FDA labeling information — Xalkori. FDA website [online], (2011).
Dancey, J. E. Epidermal growth factor receptor inhibitors in non-small cell lung cancer. Drugs 67, 1125–1138 (2007).
Park, J. W. et al. Rationale for biomarkers and surrogate end points in mechanism-driven oncology drug development. Clin. Cancer Res. 10, 3885–3896 (2004).
Gutman, S. & Kessler, L. G. The US Food and Drug Administration perspective on cancer biomarker development. Nature Rev. Cancer 6, 565–571 (2006).
Amur, S., Frueh, F. W., Lesko, L. J. & Huang, S. M. Integration and use of biomarkers in drug development, regulation and clinical practice: a US regulatory perspective. Biomark. Med. 2, 305–311 (2008).
Majewski, I. J. & Bernards, R. Taming the dragon: genomic biomarkers to individualize the treatment of cancer. Nature Med. 17, 304–312 (2011).
Dancey, J. E. et al. Guidelines for the development and incorporation of biomarker studies in early clinical trials of novel agents. Clin. Cancer Res. 16, 1745–1755 (2010).
Merlo, L. M., Pepper, J. W., Reid, B. J. & Maley, C. C. Cancer as an evolutionary and ecological process. Nature Rev. Cancer 6, 924–935 (2006).
Anderson, L. Candidate-based proteomics in the search for biomarkers of cardiovascular disease. J. Physiol. 563, 23–60 (2005).
Kelloff, G. J. et al. Progress in chemoprevention drug development: the promise of molecular biomarkers for prevention of intraepithelial neoplasia and cancer — a plan to move forward. Clin. Cancer Res. 12, 3661–3697 (2006).
Sidransky, D. Emerging molecular markers of cancer. Nature Rev. Cancer 2, 210–219 (2002).
Vogelstein, B. et al. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 319, 525–532 (1988).
Le, Q. T. & Giaccia, A. J. Therapeutic exploitation of the physiological and molecular genetic alterations in head and neck cancer. Clin. Cancer Res. 9, 4287–4295 (2003).
Ilyas, M., Straub, J., Tomlinson, I. P. & Bodmer, W. F. Genetic pathways in colorectal and other cancers. Eur. J. Cancer 35, 1986–2002 (1999).
Fearon, E. R. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990).
Dillon, D. A., Howe, C. L., Bosari, S. & Costa, J. The molecular biology of breast cancer: accelerating clinical applications. Crit. Rev. Oncog. 9, 125–140 (1998).
Leslie, N. R. & Downes, C. P. PTEN function: how normal cells control it and tumour cells lose it. Biochem. J. 382, 1–11 (2004).
Parsons, R. Human cancer, PTEN and the PI-3 kinase pathway. Semin. Cell Dev. Biol. 15, 171–176 (2004).
Sansal, I. & Sellers, W. R. The biology and clinical relevance of the PTEN tumor suppressor pathway. J. Clin. Oncol. 22, 2954–2963 (2004).
Vande Woude, G. F. et al. Reanalysis of cancer drugs: old drugs, new tricks. Clin. Cancer Res. 10, 3897–3907 (2004).
Maley, C. C. et al. Selectively advantageous mutations and hitchhikers in neoplasms: p16 lesions are selected in Barrett's esophagus. Cancer Res. 64, 3414–3427 (2004).
Reid, B. J., Blount, P. L. & Rabinovitch, P. S. Biomarkers in Barrett's esophagus. Gastrointest. Endosc. Clin. N. Am. 13, 369–397 (2003).
Polyak, K. & Garber, J. Targeting the missing links for cancer therapy. Nature Med. 17, 283–284 (2011).
Polyak, K. Molecular markers for the diagnosis and management of ductal carcinoma in situ. J. Natl Cancer Inst. Monogr. 2010, 210–213 (2010).
Peppercorn, J., Perou, C. M. & Carey, L. A. Molecular subtypes in breast cancer evaluation and management: divide and conquer. Cancer Invest. 26, 1–10 (2008).
Nielsen, T. O. et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin. Cancer Res. 16, 5222–5232 (2010).
Lenz, G. et al. Stromal gene signatures in large-B-cell lymphomas. N. Engl. J. Med. 359, 2313–2323 (2008).
Jones, S. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008).
Bozic, I. et al. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl Acad. Sci. USA 107, 18545–18550 (2010).
Parsons, D. W. et al. An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 (2008).
Salk, J. J., Fox, E. J. & Loeb, L. A. Mutational heterogeneity in human cancers: origin and consequences. Annu. Rev. Pathol. 5, 51–75 (2010).
Kelloff, G. J. et al. Perspectives on surrogate end points in the development of drugs that reduce the risk of cancer. Cancer Epidemiol. Biomarkers Prev. 9, 127–137 (2000).
Boone, C. W., Kelloff, G. J. & Freedman, L. S. Intraepithelial and postinvasive neoplasia as a stochastic continuum of clonal evolution, and its relationship to mechanisms of chemopreventive drug action. J. Cell Biochem. 53 (Suppl. 17G), 14–25 (1993).
Boone, C. W., Kelloff, G. J. & Steele, V. E. The natural history of intraepithelial neoplasia: relevance to the search for intermediate endpoint biomarkers. J. Cell Biochem. 50 (Suppl. 16G), 23–26 (1992).
Boone, C. W., Kelloff, G. J. & Steele, V. E. Natural history of intraepithelial neoplasia in humans with implications for cancer chemoprevention strategy. Cancer Res. 52, 1651–1659 (1992).
Schwarz, R. A. et al. Noninvasive evaluation of oral lesions using depth-sensitive optical spectroscopy. Cancer 115, 1669–1679 (2009).
Thekkek, N. & Richards-Kortum, R. Optical imaging for cervical cancer detection: solutions for a continuing global problem. Nature Rev. Cancer 8, 725–731 (2008).
Kelloff, G. J. et al. Progress and promise of FDG-PET imaging for cancer patient management and oncologic drug development. Clin. Cancer Res. 11, 2785–2808 (2005).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Weber, W. A. et al. Positron emission tomography in non-small-cell lung cancer: prediction of response to chemotherapy by quantitative assessment of glucose use. J. Clin. Oncol. 21, 2651–2657 (2003).
Aukema, T. S. et al. Is 18F-FDG PET/CT useful for the early prediction of histopathologic response to neoadjuvant erlotinib in patients with non-small cell lung cancer? J. Nucl. Med. 51, 1344–1348 (2010).
Zander, T. et al. Early prediction of nonprogression in advanced non-small-cell lung cancer treated with erlotinib by using [18F]fluorodeoxyglucose and [18F]fluorothymidine positron emission tomography. J. Clin. Oncol. 29, 1701–1708 (2011).
Mileshkin, L. et al. Changes in 18F-fluorodeoxyglucose and 18F-fluorodeoxythymidine positron emission tomography imaging in patients with non-small cell lung cancer treated with erlotinib. Clin. Cancer Res. 17, 3304–3315 (2011).
Galanina, N., Bossuyt, V. & Harris, L. N. Molecular predictors of response to therapy for breast cancer. Cancer J. 17, 96–103 (2011).
Tang, G. et al. Comparison of the prognostic and predictive utilities of the 21-gene Recurrence Score assay and Adjuvant! for women with node-negative, ER-positive breast cancer: results from NSABP B-14 and NSABP B-20. Breast Cancer Res. Treat. 127, 133–142 (2011).
Glas, A. M. et al. Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genomics 7, 278 (2006).
Alizadeh, A. A. et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 (2000).
Barker, A. D. et al. I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin. Pharmacol. Ther. 86, 97–100 (2009).
Slamon, D. J. et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 344, 783–792 (2001).
Pao, W. et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl Acad. Sci. USA 101, 13306–13311 (2004).
Audeh, M. W. et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet 376, 245–251 (2010).
Tutt, A. et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 376, 235–244 (2010).
Molinari, F. et al. Increased detection sensitivity for KRAS mutations enhances the prediction of anti-EGFR monoclonal antibody resistance in metastatic colorectal cancer. Clin. Cancer Res. 17, 4901–4914 (2011).
Hulka, B. S. & Wilcosky, T. Biological markers in epidemiologic research. Arch. Environ. Health 43, 83–89 (1988).
Frank, R. & Hargreaves, R. Clinical biomarkers in drug discovery and development. Nature Rev. Drug Discov. 2, 566–580 (2003).
Fearon, E. R. Human cancer syndromes: clues to the origin and nature of cancer. Science 278, 1043–1050 (1997).
Lesko, L. J. & Woodcock, J. Pharmacogenomic-guided drug development: regulatory perspective. Pharmacogenomics J. 2, 20–24 (2002).
Kelloff, G. J. & Sigman, C. C. New science-based endpoints to accelerate oncology drug development. Eur. J. Cancer 41, 491–501 (2005).
Gail, M. H. & Mai, P. L. Comparing breast cancer risk assessment models. J. Natl Cancer Inst. 102, 665–668 (2010).
Lee, S. Y. & McLeod, H. L. Pharmacogenetic tests in cancer chemotherapy: what physicians should know for clinical application. J. Pathol. 223, 15–27 (2011).
Tan, D. S. et al. Biomarker-driven early clinical trials in oncology: a paradigm shift in drug development. Cancer J. 15, 406–420 (2009).
Eisenhauer, E. A. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247 (2009).
Danila, D. C., Fleisher, M. & Scher, H. I. Circulating tumor cells as biomarkers in prostate cancer. Clin. Cancer Res. 17, 3903–3912 (2011).
Swaby, R. F. & Cristofanilli, M. Circulating tumor cells in breast cancer: a tool whose time has come of age. BMC Med. 9, 43 (2011).
Wagner, J. A., Williams, S. A. & Webster, C. J. Biomarkers and surrogate end points for fit-for-purpose development and regulatory evaluation of new drugs. Clin. Pharmacol. Ther. 81, 104–107 (2007).
Johnson, J. R. et al. Accelerated approval of oncology products: the food and drug administration experience. J. Natl Cancer Inst. 103, 636–644 (2011).
Danila, D. C., Pantel, K., Fleisher, M. & Scher, H. I. Circulating tumors cells as biomarkers: progress toward biomarker qualification. Cancer J. 17, 438–450 (2011).
Tran, L. N. et al. Comparison of treatment response classifications between unidimensional, bidimensional, and volumetric measurements of metastatic lung lesions on chest computed tomography. Acad. Radiol. 11, 1355–1360 (2004).
Zhao, B., Schwartz, L. H. & Larson, S. M. Imaging surrogates of tumor response to therapy: anatomic and functional biomarkers. J. Nucl. Med. 50, 239–249 (2009).
Beckman, R. A., Clark, J. & Chen, C. Integrating predictive biomarkers and classifiers into oncology clinical development programmes. Nature Rev. Drug Discov. 10, 735–748 (2011).
Maitournam, A. & Simon, R. On the efficiency of targeted clinical trials. Stat. Med. 24, 329–339 (2005).
Freidlin, B., McShane, L. M. & Korn, E. L. Randomized clinical trials with biomarkers: design issues. J. Natl Cancer Inst. 102, 152–160 (2010).
Freidlin, B., Jiang, W. & Simon, R. The cross-validated adaptive signature design. Clin. Cancer Res. 16, 691–698 (2010).
Berry, D. A. Bayesian clinical trials. Nature Rev. Drug Discov. 5, 27–36 (2006).
Simon, R. Validation of pharmacogenomic biomarker classifiers for treatment selection. Cancer Biomark. 2, 89–96 (2006).
Simon, R. Development and evaluation of therapeutically relevant predictive classifiers using gene expression profiling. J. Natl Cancer Inst. 98, 1169–1171 (2006).
Simon, R. Development and validation of biomarker classifiers for treatment selection. J. Stat. Plan. Inference 138, 308–320 (2008).
Hoering, A., Leblanc, M. & Crowley, J. J. Randomized Phase III clinical trial designs for targeted agents. Clin. Cancer Res. 14, 4358–4367 (2008).
Orloff, J. et al. The future of drug development: advancing clinical trial design. Nature Rev. Drug Discov. 8, 949–957 (2009).
Fukuoka, M. et al. Biomarker analyses and final overall survival results from a Phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J. Clin. Oncol. 29, 2866–2874 (2011).
Mok, T. S. et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 361, 947–957 (2009).
Temple, R. J. Enrichment designs: efficiency in development of cancer treatments. J. Clin. Oncol. 23, 4838–4839 (2005).
Clark, G. M. in Diseases of the Breast (eds Harris, J. R., Lippman, M. E., Morrow, M. & Osborne, C. K.) 489–514 (Lippincott Williams & Wilkins, Philadelphia, 2000).
Rubin, E. H., Anderson, K. M. & Gause, C. K. The BATTLE Trial: a bold step toward improving the efficiency of biomarker-based drug development. Cancer Discov. 1, 17–20 (2011).
Gold, K. A. et al. The BATTLE to personalize lung cancer prevention through reverse migration. Cancer Prev. Res. (Phila.) 4, 962–972 (2011).
Kim, E. S. et al. The BATTLE Trial: personalizing therapy for lung cancer. Cancer Discov. 1, 45–53 (2011).
Kris, M. G. et al. Identification of driver mutations in tumor speciments from 1,000 patients with lung adenocarcinoma: the NCI's Lung Cancer Mutation Consortium. J. Clin. Oncol. 29, abstract CRA7506 (2011).
Berry, D. A. Adaptive clinical trials in oncology. Nature Rev. Clin. Oncol. 8 Nov 2011 (doi: 10.1038/nrclinonc.2011.165).
Freidlin, B. & Simon, R. Adaptive signature design: an adaptive clinical trial design for generating and prospectively testing a gene expression signature for sensitive patients. Clin. Cancer Res. 11, 7872–7878 (2005).
Freidlin, B. & Korn, E. L. Biomarker-adaptive clinical trial designs. Pharmacogenomics 11, 1679–1682 (2010).
Horning, S. J. et al. Interim positron emission tomography scans in diffuse large B-cell lymphoma: an independent expert nuclear medicine evaluation of the Eastern Cooperative Oncology Group E3404 study. Blood 115, 775–777 (2010).
Seymour, L. et al. The design of Phase II clinical trials testing cancer therapeutics: consensus recommendations from the clinical trial design task force of the National Cancer Institute Investigational Drug Steering Committee. Clin. Cancer Res. 16, 1764–1769 (2010).
Ratain, M. J. & Sargent, D. J. Optimising the design of Phase II oncology trials: the importance of randomisation. Eur. J. Cancer 45, 275–280 (2009).
Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008).
Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
Chin, L., Andersen, J. N. & Futreal, P. A. Cancer genomics: from discovery science to personalized medicine. Nature Med. 17, 297–303 (2011).
Boumber, Y. & Issa, J. P. Epigenetics in cancer: what's the future? Oncology 25, 220–226, 228 (2011).
Schulman, K. A. & Tunis, S. R. A policy approach to the development of molecular diagnostic tests. Nature Biotech. 28, 1157–1159 (2010).
Lee, J. W. et al. Fit-for-purpose method development and validation for successful biomarker measurement. Pharm. Res. 23, 312–328 (2006).
Teutsch, S. M. et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Initiative: methods of the EGAPP Working Group. Genet. Med. 11, 3–14 (2009).
Pepe, M. S., Feng, Z., Janes, H., Bossuyt, P. M. & Potter, J. D. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J. Natl Cancer Inst. 100, 1432–1438 (2008).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information Box S1
Clinical study designs with biomarkers: adaptive randomization (PDF 196 kb)
Related links
Related links
FURTHER INFORMATION
FDA website — Class II Special Controls Guidance Document
FDA website — Critical Path Initiative
FDA website — Draft Guidance for Industry: Qualification Process for Drug Development Tools
FDA website — Drug–diagnostic Co-Development Concept Paper
FDA website — Guidance for Industry: Adaptive Design Clinical Trials for Drugs and Biologics
FDA website — Guidance on Pharmacogenetic Tests and Genetic Tests for Heritable Markers
FDA website — In Vitro Diagnostic Devices: Guidance for the Preparation of 510(k) Submissions
Glossary
- Adaptive clinical trial designs
-
Defined by the US Food and Drug Administration as study designs that include prospectively planned opportunities for the modification of one or more specified aspects of the study design and hypotheses based on analyses of data (usually interim data).
- Analytical and clinical validation
-
The process of assessing the biomarker assay and measuring of its performance characteristics, and determining the range of conditions (including clinical settings) under which the assay will give reproducible and accurate data.
- Biomarker qualification
-
The evidentiary process of linking a biomarker with biological processes and clinical end points. Qualification refers to the verification that the biomarker is 'fit for purpose'.
- Biomarkers
-
Factors that are objectively measured and evaluated as indicators of normal biological or pathological processes, or are pharmacological responses to therapeutic intervention.
- BRAFV600E mutation
-
A mutation in the BRAF gene leading to valine being substituted by glutamate at codon 600; found in human cancers such as papillary thyroid carcinoma, colorectal cancer, melanoma, non-small-cell lung cancer and hairy cell leukaemia.
- Clinical Laboratory Improvement Amendments
-
(CLIA). US legislation defining quality assurance practices in clinical laboratories, and requiring them to measure performance at each step of the testing process from the beginning to the test result.
- Companion diagnostics
-
In vitro diagnostic devices (assays) that provide information that is essential for the safe and effective use of a corresponding therapeutic product. The use of a companion diagnostic with a particular therapeutic product is stipulated in the instructions for use in the labelling of both the diagnostic and the corresponding therapeutic product, as well as in the labelling of any generic equivalents of the therapeutic product.
- DNA ploidy
-
A term used to describe the number of chromosome sets or deviation from the normal number of chromosomes in a cell.
- Driver mutations
-
Mutations that are causally associated with cancer development. Their presence is not required for maintenance of the final cancer (although it often is) but it must have been selected at some point during cancer development.
- Epigenetic modulation
-
Heritable changes in gene expression or cellular phenotype caused by mechanisms of interaction with the genome other than changes in the underlying DNA sequence. Examples of such changes include DNA methylation and histone deacetylation, both of which serve to suppress gene expression without altering the sequence of the silenced genes.
- Epithelial to mesenchymal transition
-
A cellular transition associated with cancer metastases in which an epithelial cell loses polarity and intercellular adhesion, and degrades basement membrane components to become a migratory mesenchymal cell.
- Fit for purpose
-
A term used in the context of biomarker qualification. A biomarker is qualified on the basis of data that support its use in specific contexts — that is, evidence linking the biomarker to specific biology and clinical end points. The biomarker is considered to be 'fit for purpose' or qualified for use in those settings in which the supporting evidence is sufficiently robust.
- Gail risk model
-
An algorithm formulated by Mitchell Gail (US National Cancer Institute) that uses personal and family history to estimate a woman's absolute risk of developing breast cancer.
- Investigational new drug application
-
An application that is submitted to a regulatory agency before a drug can be studied in humans. This application contains experimental data on: how, where and by whom the new studies will be conducted; the chemical structure of the drug; the drug's mechanism of action and metabolism; any toxic effects; and how the compound is manufactured.
- Next-generation gene sequencing
-
New technologies for high-throughput and high-speed sequencing, and potentially cost-effective sequencing of the human genome, epigenome and transcriptome (the part of the genome transcribed into RNA).
- Passenger mutations
-
Mutations that are not selected during cancer development and do not directly contribute to cancer development. Cells that acquire driver mutations already carry biologically inert somatic passenger mutations, which are included in the clonal expansion that follows and will be present in all cells of the resulting cancer.
- Standard uptake value
-
(SUV). A value that quantifies positron emission tomography imaging; calculated as a ratio of tissue radioactivity concentration at a specific time and injected dose at the time of injection, divided by body weight.
- Umbrella clinical trial
-
A clinical trial protocol into which patients are enrolled, their biomarker status is obtained and then they are assigned to treatment in one of a group of clinical trials of targeted drugs. Patients are assigned to the drug that is most likely to benefit them based on their biomarker status.
- Warburg phenomenon
-
The observation that cancer cells, unlike normal cells, preferentially utilize the glycolytic pathway rather than the Kreb's cycle to metabolize glucose.
Rights and permissions
About this article
Cite this article
Kelloff, G., Sigman, C. Cancer biomarkers: selecting the right drug for the right patient. Nat Rev Drug Discov 11, 201–214 (2012). https://doi.org/10.1038/nrd3651
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd3651
This article is cited by
-
The recent advancements in the early detection of cancer biomarkers by DNAzyme-assisted aptasensors
Journal of Nanobiotechnology (2022)
-
Antitumor pharmacological research in the era of personalized medicine
Acta Pharmacologica Sinica (2022)
-
Identification of new candidate biomarkers to support doxorubicin treatments in canine cancer patients
BMC Veterinary Research (2021)
-
Genomic data integration by WON-PARAFAC identifies interpretable factors for predicting drug-sensitivity in vivo
Nature Communications (2019)
-
Duodenojejunal Bypass Plus Sleeve Gastrectomy Reduces Infiltration of Macrophages and Secretion of TNF-α in the Visceral White Adipose Tissue of Goto-Kakizaki Rats
Obesity Surgery (2019)