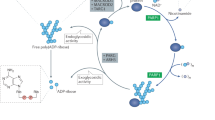
Abstract
Recent findings have thrust poly(ADP-ribose) polymerases (PARPs) into the limelight as potential chemotherapeutic targets. To provide a framework for understanding these recent observations, we review what is known about the structures and functions of the family of PARP enzymes, and then outline a series of questions that should be addressed to guide the rational development of PARP inhibitors as anticancer agents.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Chambon, P., Weill, J. D. & Mandel, P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun. 11, 39–43 (1963).
Doly, J. & Petek, F. Etude de la structure d'un composé “poly(ADP-ribose)” synthétisé par des extraits nucléaires de foie de poulet. C. R. Hebd. Seances Acad. Sci. Ser. D Sci. Nat. 263, 1341–1344 (1966).
Chambon, P., Weill, J. D., Doly, J., Strosser, M. T. & Mandel, P. On the formation of a novel adelylic compound by enzymatic extracts of liver nuclei. Biochem. Biophys. Res. Commun. 25, 638–643 (1966).
Nishizuka, Y., Ueda, K., Nakazawa, K. & Hayaishi, O. Studies on the polymer of adenosine diphosphate ribose. I. Enzymic formation from nicotinamide adenine dinuclotide in mammalian nuclei. J. Biol. Chem. 242, 3164–3171 (1967).
Sugimura, T., Fujimura, S., Hasegawa, S. & Kawamura, Y. Polymerization of the adenosine 5′-diphosphate ribose moiety of NAD by rat liver nuclear enzyme. Biochim. Biophys. Acta 138, 438–441 (1967).
Yamada, M., Miwa, M. & Sugimura, T. Studies on poly (adenosine diphosphate-ribose): X. Properties of a partially purified poly (adenosine diphosphate-ribose) polymerase. Arch. Biochem. Biophys. 146, 579–586 (1971).
Okayama, H., Edson, C. M., Fukushima, M., Ueda, K. & Hayaishi, O. Purification and properties of poly(adenosine diphosphate ribose) synthetase. J. Biol. Chem. 252, 7000–7005 (1977).
Juarez-Salinas, H., Sims, J. L. & Jacobson, M. K. Poly(ADP-ribose) levels in carcinogen-treated cells. Nature 282, 740–741 (1979).
Benjamin, R. C. & Gill, D. M. ADP-ribosylation in mammalian cell ghosts. Dependence of poly(ADP-ribose) synthesis on strand breakage in DNA. J. Biol. Chem. 255, 10493–10501 (1980).
Durkacz, B. W., Omidiji, O., Gray, D. A. & Shall, S. (ADP-ribose)n participates in DNA excision repair. Nature 283, 593–596 (1980).
Shall, S. & de Murcia, G. Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat. Res. 460, 1–15 (2000).
Amé, J. C. et al. PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 274, 17860–17868 (1999).
Berger, S. J., Sudar, D. C. & Berger, N. A. Metabolic consequences of DNA damage: DNA damage induces alterations in glucose metabolism by activation of poly (ADP-ribose) polymerase. Biochem. Biophys. Res. Commun. 134, 227–232 (1986).
Carson, D. A., Carrera, C. J., Wasson, D. B. & Yamanaka, H. Programmed cell death and adenine deoxynucleotide metabolism in human lymphocytes. Adv. Enzyme Regul. 27, 395–404 (1988).
David, K. K., Andrabi, S. A., Dawson, T. M. & Dawson, V. L. Parthanatos, a messenger of death. Front. Biosci. 14, 1116–1128 (2009).
Bryant, H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005).
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005).
Fong, P. C. et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 361, 123–134 (2009).
Gradwohl, G. et al. The second zinc-finger domain of poly(ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proc. Natl Acad. Sci. USA 87, 2990–2994 (1990).
Hassa, P. O. & Hottiger, M. O. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 13, 3046–3082 (2008).
Langelier, M. F., Servent, K. M., Rogers, E. E. & Pascal, J. M. A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J. Biol. Chem. 283, 4105–4114 (2008).
Tao, Z., Gao, P., Hoffman, D. W. & Liu, H. W. Domain C of human poly(ADP-ribose) polymerase-1 is important for enzyme activity and contains a novel zinc-ribbon motif. Biochemistry 47, 5804–5813 (2008).
Altmeyer, M., Messner, S., Hassa, P. O., Fey, M. & Hottiger, M. O. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 37, 3723–3738 (2009).
Tao, Z., Gao, P. & Liu, H. W. Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J. Am. Chem. Soc. 131, 14258–14260 (2009).
Kameshita, I., Matsuda, Z., Taniguchi, T. & Shizuta, Y. Poly (ADP-ribose) synthetase. Separation and identification of three proteolytic fragments as the substrate-binding domain, the DNA-binding domain, and the automodification domain. J. Biol. Chem. 259, 4770–4776 (1984).
Haince, J. F. et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 283, 1197–1208 (2008).
D'Amours, D., Desnoyers, S., D'Silva, I. & Poirier, G. G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342, 249–268 (1999).
Gagné, J. P. et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 36, 6959–6976 (2008).
Timinszky, G. et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nature Struct. Mol. Biol. 16, 923–929 (2009).
Kraus, W. L. New functions for an ancient domain. Nature Struct. Mol. Biol. 16, 904–907 (2009).
Ahel, D. et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325, 1240–1243 (2009).
Gottschalk, A. J. et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl Acad. Sci. USA 106, 13770–13774 (2009).
Masson, M. et al. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell Biol. 18, 3563–3571 (1998).
El-Khamisy, S. F., Masutani, M., Suzuki, H. & Caldecott, K. W. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 31, 5526–5533 (2003).
Ogata, N., Ueda, K., Kagamiyama, H. & Hayaishi, O. ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J. Biol. Chem. 255, 7616–7620 (1980).
Poirier, G. G., de Murcia, G., Jongstra-Bilen, J., Niedergang, C. & Mandel, P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl Acad. Sci. USA 79, 3423–3427 (1982).
Tulin, A. & Spradling, A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 299, 560–562 (2003).
Satoh, M. S. & Lindahl, T. Role of poly(ADP-ribose) formation in DNA repair. Nature 356, 356–358 (1992).
Zahradka, P. & Ebisuzaki, K. A shuttle mechanism for DNA–protein interactions. The regulation of poly(ADP-ribose) polymerase. Eur. J. Biochem. 127, 579–585 (1982).
Meyer-Ficca, M. L., Meyer, R. G., Coyle, D. L., Jacobson, E. L. & Jacobson, M. K. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp. Cell Res. 297, 521–532 (2004).
Oka, S., Kato, J. & Moss, J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 281, 705–713 (2006).
Oka, J., Ueda, K., Hayaishi, O., Komura, H. & Nakanishi, K. ADP-ribosyl protein lyase. Purification, properties, and identification of the product. J. Biol. Chem. 259, 986–995 (1984).
Okayama, H., Honda, M. & Hayaishi, O. Novel enzyme from rat liver that cleaves an ADP-ribosyl histone linkage. Proc. Natl Acad. Sci. USA 75, 2254–2257 (1978).
Hardie, D. G. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nature Rev. Mol. Cell Biol. 8, 774–785 (2007).
Huang, Q., Wu, Y. T., Tan, H. L., Ong, C. N. & Shen, H. M. A novel function of poly(ADP-ribose) polymerase-1 in modulation of autophagy and necrosis under oxidative stress. Cell Death Differ. 16, 264–277 (2009).
Haince, J. F. et al. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J. Biol. Chem. 282, 16441–16453 (2007).
Yang, Y. G., Cortes, U., Patnaik, S., Jasin, M. & Wang, Z. Q. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene 23, 3872–3882 (2004).
Sugimura, K., Takebayashi, S., Taguchi, H., Takeda, S. & Okumura, K. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J. Cell Biol. 183, 1203–1212 (2008).
Bryant, H. E. et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 28, 2601–2615 (2009).
Audebert, M., Salles, B. & Calsou, P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 279, 55117–55126 (2004).
Veuger, S. J., Curtin, N. J., Smith, G. C. & Durkacz, B. W. Effects of novel inhibitors of poly(ADP-ribose) polymerase-1 and the DNA-dependent protein kinase on enzyme activities and DNA repair. Oncogene 23, 7322–7329 (2004).
Wang, M. et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 34, 6170–6182 (2006).
Caiafa, P., Guastafierro, T. & Zampieri, M. Epigenetics: poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns. FASEB J. 23, 672–678 (2009).
Kraus, W. L. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 20, 294–302 (2008).
Krishnakumar, R. et al. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319, 819–821 (2008).
Simbulan-Rosenthal, C. M. et al. Misregulation of gene expression in primary fibroblasts lacking poly(ADP-ribose) polymerase. Proc. Natl Acad. Sci. USA 97, 11274–11279 (2000).
Frizzell, K. M. et al. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J. Biol. Chem. 284, 33926–33938 (2009).
Amé, J. C., Spenlehauer, C. & de Murcia, G. The PARP superfamily. Bioessays 26, 882–893 (2004).
Kleine, H. et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell 32, 57–69 (2008).
Karlberg, T., Hammarstrom, M., Schutz, P., Svensson, L. & Schuler, H. Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888. Biochemistry 49, 1056–1058 (2010).
Lehtio, L. et al. Zinc binding catalytic domain of human tankyrase 1. J. Mol. Biol. 379, 136–145 (2008).
Lehtio, L. et al. Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3. J. Med. Chem. 52, 3108–3111 (2009).
Ménissier de Murcia, J. et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 22, 2255–2263 (2003).
Chiang, Y. J. et al. Tankyrase 1 and tankyrase 2 are essential but redundant for mouse embryonic development. PLoS ONE 3, e2639 (2008).
Yelamos, J., Schreiber, V. & Dantzer, F. Toward specific functions of poly(ADP-ribose) polymerase-2. Trends Mol. Med. 14, 169–178 (2008).
Shieh, W. M. et al. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem. 273, 30069–30072 (1998).
Sallmann, F. R., Vodenicharov, M. D., Wang, Z. Q. & Poirier, G. G. Characterization of sPARP-1. An alternative product of PARP-1 gene with poly(ADP-ribose) polymerase activity independent of DNA strand breaks. J. Biol. Chem. 275, 15504–15511 (2000).
Zong, W. X., Ditsworth, D., Bauer, D. E., Wang, Z. Q. & Thompson, C. B. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 18, 1272–1282 (2004).
Liu, Y. et al. Vault poly(ADP-ribose) polymerase is associated with mammalian telomerase and is dispensable for telomerase function and vault structure in vivo. Mol. Cell. Biol. 24, 5314–5323 (2004).
Raval-Fernandes, S., Kickhoefer, V. A., Kitchen, C. & Rome, L. H. Increased susceptibility of vault poly(ADP-ribose) polymerase-deficient mice to carcinogen-induced tumorigenesis. Cancer Res. 65, 8846–8852 (2005).
Hsiao, S. J. & Smith, S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie 90, 83–92 (2008).
Sbodio, J. I. & Chi, N. W. Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1. NuMA contains this RXXPDG motif and is a novel tankyrase partner. J. Biol. Chem. 277, 31887–31892 (2002).
Canudas, S. et al. Protein requirements for sister telomere association in human cells. EMBO J. 26, 4867–4878 (2007).
Hsiao, S. J. & Smith, S. Sister telomeres rendered dysfunctional by persistent cohesion are fused by NHEJ. J. Cell Biol. 184, 515–526 (2009).
Huang, S. M. et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 (2009).
Nobori, T., Yamanaka, H. & Carson, D. A. Poly(ADP-ribose) polymerase inhibits DNA synthesis initiation in the absence of NAD. Biochem. Biophys. Res. Commun. 163, 1113–1118 (1989).
Zaremba, T. & Curtin, N. J. PARP inhibitor development for systemic cancer targeting. Anticancer Agents Med. Chem. 7, 515–523 (2007).
Satoh, M. S., Poirier, G. G. & Lindahl, T. Dual function for poly(ADP-ribose) synthesis in response to DNA strand breakage. Biochemistry 33, 7099–7106 (1994).
Farzaneh, F., Zalin, R., Brill, D. & Shall, S. DNA strand breaks and ADP-ribosyl transferase activation during cell differentiation. Nature 300, 362–366 (1982).
Johnstone, A. P. & Williams, G. T. Role of DNA breaks and ADP-ribosyl transferase activity in eukaryotic differentiation demonstrated in human lymphocytes. Nature 300, 368–370 (1982).
Saleh-Gohari, N. et al. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell Biol. 25, 7158–7169 (2005).
Anders, C. & Carey, L. A. Understanding and treating triple-negative breast cancer. Oncology 22, 1233–1239 (2008).
Turner, N., Tutt, A. & Ashworth, A. Hallmarks of 'BRCAness' in sporadic cancers. Nature Rev. Cancer 4, 814–819 (2004).
McCabe, N. et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 66, 8109–8115 (2006).
Mendes-Pereira, A. M. et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol. Med. 1, 315–322 (2009).
Ashworth, A. Drug resistance caused by reversion mutation. Cancer Res. 68, 10021–10023 (2008).
Sakai, W. et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451, 1116–1120 (2008).
Edwards, S. L. et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451, 1111–1115 (2008).
Ratnam, K. & Low, J. A. Current development of clinical inhibitors of poly(ADP-ribose) polymerase in oncology. Clin. Cancer Res. 13, 1383–1388 (2007).
Plummer, R. et al. Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with temozolomide in patients with advanced solid tumors. Clin. Cancer Res. 14, 7917–7923 (2008).
O'Shaughnessy, J. et al. Efficacy of BSI-201, a poly (ADP-ribose) polymerase-1 (PARP1) inhibitor, in combination with gemcitabine/carboplatin (G/C) in patients with metastatic triple-negative breast cancer (TNBC): results of a randomized Phase II trial. J. Clin. Oncol. 27, 3 (2009).
Ossovskaya, V. et al. BSI-201 enhances the activity of multiple classes of cytotoxic agents and irradiation in triple negative breast cancer. Abstract 5552. Proc. Annu. Meet. Am. Assoc. Cancer Res. (2009).
Pacher, P. & Szabo, C. Role of poly(ADP-ribose) polymerase 1 (PARP-1) in cardiovascular diseases: the therapeutic potential of PARP inhibitors. Cardiovasc. Drug Rev. 25, 235–260 (2007).
Goldberg, S., Visochek, L., Giladi, E., Gozes, I. & Cohen-Armon, M. PolyADP-ribosylation is required for long-term memory formation in mammals. J. Neurochem. 111, 72–79 (2009).
Morrison, C. et al. Genetic interaction between PARP and DNA-PK in V(D)J. recombination and tumorigenesis. Nature Genet. 17, 479–482 (1997).
Tong, W. M. et al. Synergistic role of Ku80 and poly(ADP-ribose) polymerase in suppressing chromosomal aberrations and liver cancer formation. Cancer Res. 62, 6990–6996 (2002).
Tong, W. M. et al. Null mutation of DNA strand break-binding molecule poly(ADP-ribose) polymerase causes medulloblastomas in p53−/− mice. Am. J. Pathol. 162, 343–352 (2003).
Lilyestrom, W., van der Woerd, M. J., Clark, N. & Luger, K. Structural and biophysical studies of human PARP-1 in complex with damaged DNA. J. Mol. Biol. 395, 983–994 (2010).
Formentini, L. et al. Poly(ADP-ribose) catabolism triggers AMP-dependent mitochondrial energy failure. J. Biol. Chem. 284, 17668–17676 (2009).
Loseva, O. et al. Poly(ADP-ribose) polymerase-3 (PARP-3) is a mono-ADP ribosylase that activates PARP-1 in absence of DNA. J. Biol. Chem. 11 Jan 2010 (doi: 10.1074/jbc.M109.077834).
McCabe, N. et al. Targeting tankyrase 1 as a therapeutic strategy for BRCA-associated cancer. Oncogene 28, 1465–1470 (2009).
Turner, N. C. et al. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J. 27, 1368–1377 (2008).
Mendeleyev, J., Kirsten, E., Hakam, A., Buki, K. G. & Kun, E. Potential chemotherapeutic activity of 4-iodo-3-nitrobenzamide. Metabolic reduction to the 3-nitroso derivative and induction of cell death in tumor cells in culture. Biochem. Pharmacol. 50, 705–714 (1995).
Moore, J. et al. Treatment of cancer. US Patent Application Publication US2008/0103104 A1 (2008).
Konishi, Y. et al. Possible model of liver carcinogenesis using inhibitors of NAD+ ADP ribosyl transferase in rats. Toxicol. Pathol. 14, 483–488 (1986).
Takahashi, S. et al. Enhancement of DEN initiation of liver carcinogenesis by inhibitors of NAD+ ADP ribosyl transferase in rats. Carcinogenesis 5, 901–906 (1984).
Andrabi, S. A. et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl Acad. Sci. USA 103, 18308–18313 (2006).
McLennan, A. G. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 63, 123–143 (2006).
Huang, Q. & Shen, H. M. To die or to live: the dual role of poly(ADP-ribose) polymerase-1 in autophagy and necrosis under oxidative stress and DNA damage. Autophagy 5, 273–276 (2009).
Munoz-Gamez, J. A. et al. PARP-1 is involved in autophagy induced by DNA damage. Autophagy 5, 61–74 (2009).
Otto, H. et al. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs). BMC Genomics 6, 139 (2005).
Alvarez-Gonzalez, R. & Jacobson, M. K. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry 26, 3218–3224 (1987).
Rippmann, J. F., Damm, K. & Schnapp, A. Functional characterization of the poly(ADP-ribose) polymerase activity of tankyrase 1, a potential regulator of telomere length. J. Mol. Biol. 323, 217–224 (2002).
Sbodio, J. I., Lodish, H. F. & Chi, N. W. Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase). Biochem. J. 361, 451–459 (2002).
Augustin, A. et al. PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. J. Cell Sci. 116, 1551–1562 (2003).
Bauer, P. I., Buki, K. G., Hakam, A. & Kun, E. Macromolecular association of ADP-ribosyltransferase and its correlation with enzymic activity. Biochem. J. 270, 17–26 (1990).
Mendoza-Alvarez, H. & Alvarez-Gonzalez, R. Poly(ADP-ribose) polymerase is a catalytic dimer and the automodification reaction is intermolecular. J. Biol. Chem. 268, 22575–22580 (1993).
de Murcia, J. M. et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl Acad. Sci. USA 94, 7303–7307 (1997).
Masutani, M. et al. Function of poly(ADP-ribose) polymerase in response to DNA damage: gene-disruption study in mice. Mol. Cell Biochem. 193, 149–152 (1999).
Wang, Z. Q. et al. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 9, 509–520 (1995).
Yelamos, J. et al. PARP-2 deficiency affects the survival of CD4+CD8+ double-positive thymocytes. EMBO J. 25, 4350–4360 (2006).
Yeh, T. Y. et al. Hypermetabolism, hyperphagia, and reduced adiposity in tankyrase-deficient mice. Diabetes 58, 2476–2485 (2009).
Chiang, Y. J. et al. Generation and characterization of telomere length maintenance in tankyrase 2-deficient mice. Mol. Cell. Biol. 26, 2037–2043 (2006).
Hsiao, S. J., Poitras, M. F., Cook, B. D., Liu, Y. & Smith, S. Tankyrase 2 poly(ADP-ribose) polymerase domain-deleted mice exhibit growth defects but have normal telomere length and capping. Mol. Cell. Biol. 26, 2044–2054 (2006).
Albert, J. M. et al. Inhibition of poly(ADP-ribose) polymerase enhances cell death and improves tumor growth delay in irradiated lung cancer models. Clin. Cancer Res. 13, 3033–3042 (2007).
Clarke, M. J. et al. Effective sensitization of temozolomide by ABT-888 is lost with development of temozolomide resistance in glioblastoma xenograft lines. Mol. Cancer Ther. 8, 407–414 (2009).
Donawho, C. K. et al. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin. Cancer Res. 13, 2728–2737 (2007).
Horton, T. M. et al. Poly(ADP-ribose) polymerase inhibitor ABT-888 potentiates the cytotoxic activity of temozolomide in leukemia cells: influence of mismatch repair status and O6-methylguanine-DNA methyltransferase activity. Mol. Cancer Ther. 8, 2232–2242 (2009).
Liu, S. K. et al. A novel poly(ADP-ribose) polymerase inhibitor, ABT-888, radiosensitizes malignant human cell lines under hypoxia. Radiother Oncol. 88, 258–268 (2008).
Liu, X. et al. Potentiation of temozolomide cytotoxicity by poly(ADP)ribose polymerase inhibitor ABT-888 requires a conversion of single-stranded DNA damages to double-stranded DNA breaks. Mol. Cancer Res. 6, 1621–1629 (2008).
Penning, T. D. et al. Discovery of the poly(ADP-ribose) polymerase (PARP) inhibitor 2-[(R.)-2-methylpyrrolidin-2-yl]- 1H-benzimidazole-4-carboxamide (ABT-888) for the treatment of cancer. J. Med. Chem. 52, 514–523 (2009).
Daniel, R. A. et al. Inhibition of poly(ADP-ribose) polymerase-1 enhances temozolomide and topotecan activity against childhood neuroblastoma. Clin. Cancer Res. 15, 1241–1249 (2009).
Thomas, H. D. et al. Preclinical selection of a novel poly(ADP-ribose) polymerase inhibitor for clinical trial. Mol. Cancer Ther. 6, 945–956 (2007).
Dungey, F. A., Caldecott, K. W. & Chalmers, A. J. Enhanced radiosensitization of human glioma cells by combining inhibition of poly(ADP-ribose) polymerase with inhibition of heat shock protein 90. Mol. Cancer Ther. 8, 2243–2254 (2009).
Dungey, F. A., Loser, D. A. & Chalmers, A. J. Replication-dependent radiosensitization of human glioma cells by inhibition of poly(ADP-ribose) polymerase: mechanisms and therapeutic potential. Int. J. Radiat. Oncol. Biol. Phys. 72, 1188–1197 (2008).
Evers, B. et al. Selective inhibition of BRCA2-deficient mammary tumor cell growth by AZD2281 and cisplatin. Clin. Cancer Res. 14, 3916–3925 (2008).
Hay, T. et al. Poly(ADP-ribose) polymerase-1 inhibitor treatment regresses autochthonous Brca2/p53-mutant mammary tumors in vivo and delays tumor relapse in combination with carboplatin. Cancer Res. 69, 3850–3855 (2009).
Menear, K. A. et al. 4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phth alazin-1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J. Med. Chem. 51, 6581–6591 (2008).
Rottenberg, S. et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl Acad. Sci. USA 105, 17079–17084 (2008).
Miknyoczki, S. et al. The selective poly(ADP-ribose) polymerase-1(2) inhibitor, CEP-8983, increases the sensitivity of chemoresistant tumor cells to temozolomide and irinotecan but does not potentiate myelotoxicity. Mol. Cancer Ther. 6, 2290–2302 (2007).
Jones, P. et al. Discovery of 2-{4-[(3S)-Piperidin-3-yl]phenyl}-2H-indazole-7-.carboxamide (MK-4827): a novel oral poly(ADP-ribose)polymerase (PARP) inhibitor+ efficacious in BRCA-1 and -2 mutant tumors. J. Med. Chem. 52, 7170–7185 (2009).
Acknowledgements
The authors are supported by research funds from a Canada research chair in proteomics, the Canadian Institutes of Health Research (CIHR grants MOP-74648 and IG1-14052), the Cancer Research Society, the Alberta Cancer Board and the National Institutes of Health (NIH grant P50 CA136393-01).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Rights and permissions
About this article
Cite this article
Rouleau, M., Patel, A., Hendzel, M. et al. PARP inhibition: PARP1 and beyond. Nat Rev Cancer 10, 293–301 (2010). https://doi.org/10.1038/nrc2812
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc2812
This article is cited by
-
Discovery of novel 2,3,4,5-tetrahydrospiro[benzo[c]azepine-1,1’-cyclohexan]-5-ol derivatives as PARP-1 inhibitors
BMC Chemistry (2023)
-
Targetable lesions and proteomes predict therapy sensitivity through disease evolution in pediatric acute lymphoblastic leukemia
Nature Communications (2023)
-
Systematic profiling of conditional pathway activation identifies context-dependent synthetic lethalities
Nature Genetics (2023)
-
Preclinical evaluation of a brain penetrant PARP PET imaging probe in rat glioblastoma and nonhuman primates
European Journal of Nuclear Medicine and Molecular Imaging (2023)
-
Triple negative breast cancer: approved treatment options and their mechanisms of action
Journal of Cancer Research and Clinical Oncology (2023)