Abstract
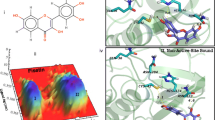
Herbicides targeting grass plastidic acetyl-CoA carboxylase (ACCase, EC 6.4.1.2) are selectively effective against graminicides. The intensive worldwide use of this herbicide family has selected for resistance genes in a number of grass weed species. Recently, the active-site W374C mutation was found to confer multi-drug resistance toward haloxyfop (HF), fenoxaprop (FR), Diclofop (DF), and clodinafop (CF) in A. myosuroides. In order to uncover the resistance mechanism due to W374C mutation, the binding of above-mentioned four herbicides to both wild-type and the mutant-type ACCase was investigated in the current work by molecular docking and molecular dynamics (MD) simulations. The binding free energies were calculated by molecular mechanics-Poisson-Boltzmann surface area (MM/PBSA) method. The calculated binding free energy values for four herbicides were qualitatively consistent with the experimental order of IC50 values. All the computational model and energetic results indicated that the W374C mutation has great effects on the conformational change of the binding pocket and the ligand-protein interactions. The most significant conformational change was found to be associated with the aromatic amino acid residues, such as Phe377, Tyr161′ and Trp346. As a result, the π-π interaction between the ligand and the residue of Phe377 and Tyr161′, which make important contributions to the binding affinity, was decreased after mutation and the binding affinity for the inhibitors to the mutant-type ACCase was less than that to the wild-type enzyme, which accounts for the molecular basis of herbicidal resistance. The structural role and mechanistic insights obtained from computational simulations will provide a new starting point for the rational design of novel inhibitors to overcome drug resistance associated with W374C mutation.





Similar content being viewed by others
References
Nikolau BJ, Ohlrogge JB, Wurtele ES (2003) Plant biotin-containing carboxylases. Arch Biochem Biophy 414:211–222
Egli MA, Gengenbach BG, Gronwald JW, Somers DA, Wyse DL (1993) Characterization of maize acetyl-coenzyme A carboxylase. Plant Physiol 101:499–506
Harwood JL (1988) Fatty acid metabolism. Annu Rev Plant Physiol 39:101–138
Post-Beittenmiller D (1996) Biochemistry and molecular biology of wax production in plants. Annu Rev Plant Physiol Plant Mol Biol 47:405–430
Zhang H, Tweet B, Tong L (2004) Molecular basis for the inhibiton of the carboxyltransferase domain of acetyl coenzyme-A carboxylase by haloxyfop and diclofop. Proc Natl Acad Sci USA 101:5910–5915
Hofer U, Muehlebach M, Hole S, Zoschke A (2006) Pinoxaden-for broad spectrum grass weed management in ceral crops. J Plant Dis Prot 20:989–995
Rendina AR, Craig-Kennard AC, Beaudoin JD, Breen MK (1990) Inhibition of acetyl-coenzyme A carboxylase by two classes of grass-selective herbicides. J Agric Food Chem 38:1282–1287
Burton JD, Gronwald JW, Keith RA, Somers DA, Gegenbach BG, Wyse DL (1991) Kinetics of inhibition of acetyl-coenzyme A carboxylase by sethoxydim and haloxyfop. Pestic Biochem Physiol 39:100–109
Yu Q, Collavo C, Zheng MQ, Owen M, Sattin M, Powles S (2007) Diversity of ACCase mutations in resistant Lolium populations: evaluation using clethodim. Plant Physiol 145:547–558
Délye C, Zhang XQ, Michel S, Matejicek A, Powles SB (2005) Molecular bases for sensitivity to acetyl-coenzyme a carboxylase inhibitors in black-grass. Plant Physiol 137:794–806
Liu WJ, Harrison DK, Chalupska D, Gornicki P, O’Donnell CC, Adkins SW, Haselkorn R, Williams RR (2007) Single-site mutations in the carboxyltransferase domain of plastid acetyl-CoA carboxylase confer resistance to grass-specific herbicides. Proc Natl Acad Sci USA 104:3627–3632
Délye C (2005) Weed resistance to acetyl coenzyme A carboxylase inhibitors: an update. Weed Sci 53:728–746
Zhu XL, Hao GF, Zhan CG, Yang GF (2009) Computational simulations of the interactions between acetyl-coenzyme-a carboxylase and clodinafop: resistance mechanism due to active and nonactive site mutations. J Chem Inf Model 49:1936–1943
Hao GF, Zhu XL, Ji FQ, Zhang L, Yang GF, Zhan CG (2009) Understanding mechanism of drug resistance due to a codon deletion in protoporphrinogen oxidase through computational modeling. J Phys Chem B 113:4865–4875
Rafi SB, Cui GL, Song K, Cheng XL, Tonge PJ, Simmerling C (2006) Insight through molecular mechanics poisson−boltzmann surface area calculations into the binding affinity of triclosan and three analogues for fabi, the e. coli enoyl reductase. J Med Chem 49:4574–4580
Hou TJ, Yu R (2007) Molecular dynamics and free energy studies on the wild-type and double mutant hiv-1 protease complexed with amprenavir and two amprenavir-related inhibitors: mechanism for binding and drug resistance. J Med Chem 50:1177–1188
Weis A, Katebzadeh K, Söderhjelm P, Nilsson I, Ryde U (2006) Ligand affinities predicted with the mm/pbsa method: dependence on the simulation method and the force field. J Med Chem 49:6596–6606
SYBYL 7.1, Tripos International, 1699 South Hanley Rd, St Louis, Missouri, 63144, USA
Zhu XL, Zhang L, Chen Q, Wan J, Yang GF (2006) Interactions of aryloxyphen- oxypropionic acids with sensitive and resistant acetyl-coenzyme a carboxylase by homology modeling and molecular dynamic simulations. J Chem Inf Model 46:1819–1826
Gao DQ, Cho H, Yang WC, Pan YM, Yang GF, Tai HH, Zhan CG (2006) Computational design of a human butyrylcholinesterase mutant for accelerating cocaine hydrolysis based on the transition-state simulation. Angew Chem Int Ed 45:653–657
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65:712–725
Case, DA, Darden T, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R (2004) AMBER8 users’ manual; University of California
Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman J, Hornak V, Cui G, Beroza P, Schafmeister C, Caldwell JW, Rose WS, Kollman PA (2004) AMBER8. University of California, San Francisco, CA
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25:1157–1174
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA Jr, Verven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokumo K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PM, Wong MW, Gonzalez C, Pople JA (2003) Gaussian 03, Rev A.1. Gaussian Inc, Pittsburgh
Cornell WD, Cieplak P, Bayly CI, Kollmann PA (1993) Application of RESP charges to calculate conformational energies, hydrogen bond energies, and free energies of solvation. J Am Chem Soc 115:9620–9631
Jorgensen WL, Chandrasekhar J, Madura JD (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: An N· log (N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Essmann U, Perera L, Berkowitz ML (1995) A smooth particle mesh Ewald method. J Chem Phys 103:8577–8593
Srinivasan J, Cheatham TE III, Kollman P, Case DA (1998) Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate-DNA helices. J Am Chem Soc 120:9401–9409
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee T, Duan Y, Wang W, Donini O, Ciplak P, Srinivasan J, Case DA, Cheatham TE III (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continum models. Acc Chem Res 33:889–897
Sitkoff D, Sharp KA, Honig B (1994) Accurate calculation of hydration free energies using macroscopic solvent models. J Phys Chem 98:1978–1988
Gilson MK, Sharp KA, Honig BH (1987) Calculating the electrostatic potential of molecules in solution: method and error assessment. J Comput Chem 9:327–335
Ji FQ, Niu CW, Chen CN, Chen Q, Yang GF, Xi Z, Zhan CG (2008) Computational design and discovery of conformationally flexible inhibitors of acetohydroxyacid synthase to overcome drug resistance associated with the W586L mutation. Chem Med Chem 3:1203–1206
Pan YM, Gao DQ, Zhan CG (2008) Modeling the catalysis of anti-cocaine catalytic antibody: competing reaction pathways and free energy barriers. J Am Chem Soc 130:5140–5149
Acknowledgments
The research was supported in part by the National Basic Research Program of China (No. 2010CB126103) and the National Nature Science Foundation of China (No. 20925206 20932005, and 20902034).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 172 kb)
Rights and permissions
About this article
Cite this article
Zhu, XL., Yang, WC., Yu, NX. et al. Computational simulations of structural role of the active-site W374C mutation of acetyl-coenzyme-A carboxylase: Multi-drug resistance mechanism. J Mol Model 17, 495–503 (2011). https://doi.org/10.1007/s00894-010-0742-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-010-0742-4