Abstract
The world’s first 194-cm-long total-body PET/CT scanner (uEXPLORER) has been built by the EXPLORER Consortium to offer a transformative platform for human molecular imaging in clinical research and health care. Its total-body coverage and ultra-high sensitivity provide opportunities for more accurate tracer kinetic analysis in studies of physiology, biochemistry, and pharmacology. The objective of this study was to demonstrate the capability of total-body parametric imaging and to quantify the improvement in image quality and kinetic parameter estimation by direct and kernel reconstruction of the uEXPLORER data. Methods: We developed quantitative parametric image reconstruction methods for kinetic analysis and used them to analyze the first human dynamic total-body PET study. A healthy female subject was recruited, and a 1-h dynamic scan was acquired during and after an intravenous injection of 256 MBq of 18F-FDG. Dynamic data were reconstructed using a 3-dimensional time-of-flight list-mode ordered-subsets expectation maximization (OSEM) algorithm and a kernel-based algorithm with all quantitative corrections implemented in the forward model. The Patlak graphical model was used to analyze the 18F-FDG kinetics in the whole body. The input function was extracted from a region over the descending aorta. For comparison, indirect Patlak analysis from reconstructed frames and direct reconstruction of parametric images from the list-mode data were obtained for the last 30 min of data. Results: Images reconstructed by OSEM showed good quality with low noise, even for the 1-s frames. The image quality was further improved using the kernel method. Total-body Patlak parametric images were obtained using either indirect estimation or direct reconstruction. The direct reconstruction method improved the parametric image quality, having a better contrast-versus-noise tradeoff than the indirect method, with a 2- to 3-fold variance reduction. The kernel-based indirect Patlak method offered image quality similar to the direct Patlak method, with less computation time and faster convergence. Conclusion: This study demonstrated the capability of total-body parametric imaging using the uEXPLORER. Furthermore, the results showed the benefits of kernel-regularized reconstruction and direct parametric reconstruction. Both can achieve superior image quality for tracer kinetic studies compared with the conventional indirect OSEM for total-body imaging.
PET has been widely applied in oncology, neurology, and cardiology by providing sensitive molecular imaging of biologic processes using radiotracers (1). By means of dynamic acquisition over time, PET can measure the spatiotemporal distribution of radiotracers in vivo, which can be used to provide assays of physiologically and biologically relevant information such as cerebral blood volume, blood flow, glucose metabolism, oxygen use, DNA synthesis, signal transduction, immune system activation, cancer cell phenotyping for molecularly targeted therapies, pharmacokinetics, and pharmacodynamics, through mathematic modeling of the tracer kinetics (2). Because of the limited length of current clinical PET scanners (e.g., typically covering an axial field of view [AFOV] of 15–30 cm), most dynamic PET scans in the past have been restricted to a single bed position over a limited region of the human body at one time. To acquire whole-body dynamic imaging (e.g., typically from head or neck to upper thigh), multiple-bed-position scans (step-and-shoot mode or continuous bed motion) with a sequential multipass protocol were introduced (3,4). However, two major drawbacks exist in this approach: first, the temporal sampling resolution is limited by the multiple-bed-position acquisition, and fast tracer dynamics cannot be captured except in a single body region; second, the scan duration is divided among multiple bed positions, resulting in reduced sensitivity and a low signal-to-noise ratio for parametric imaging.
To overcome the limitation of low sensitivity and short AFOV in current clinical PET scanners, the EXPLORER Consortium has been developing very large AFOV imaging systems to offer a transformative platform for biomedical research and clinical applications (5). To date, the consortium has built a 45-cm-long photomultiplier tube–based PET system for nonhuman primate imaging (mini-EXPLORER-I) (6), a 48-cm-long silicon photomultiplier–based PET system for veterinary medical imaging (mini-EXPLORER-II) (7), a scalable 70-cm-long digital silicon photomultiplier PET system for torso imaging (PennPET EXPLORER) (8), and a recently completed 194-cm-long PET/CT system for total-body imaging (uEXPLORER is the product name of the 194-cm-long PET/CT device built by United Imaging Healthcare, and we use this name to refer to the specific scanner tested in this work) (9). Compared with conventional scanners, the massively increased sensitivity of the uEXPLORER improves PET image quality through a high signal-to-noise ratio that supports high spatial and temporal resolution and is also predicted to provide much better lesion detection and region-of-interest (ROI) quantification (10,11). Its entire-body coverage and ultra-high sensitivity provide opportunities for more accurate tracer kinetic analysis in studies of human physiology, biochemistry, and pharmacology. The uEXPLORER provides the first opportunity to obtain dynamic imaging of the entire body of an adult human with simultaneous coverage of all organs, tissues, and cells. The blood input function can also be extracted from the aorta or left ventricle, thus alleviating the need for invasive blood sampling, provided that metabolite activity is negligible over the time of the imaging study. The high sensitivity of the uEXPLORER also permits high-quality parametric imaging on the voxel level for the whole body, which can better model heterogeneous regional tracer kinetics than ROI-based analysis.
Parametric image reconstruction methods have been developed for linear graphical models (e.g., Patlak plot and Logan plot) and compartmental models (12). The conventional approach is to reconstruct the dynamic temporal frames first and then to fit the time–activity curves of each voxel with a kinetic model. This approach is commonly referred to as the indirect method. It is easy to implement but usually results in high noise in the parametric images because the noise distribution in reconstructed images is difficult to model (13). Penalized maximum-likelihood image reconstruction methods can be used to improve the indirect parametric images (14). Regularization using kernel-based methods has also been developed to enhance the dynamic PET reconstruction by incorporating anatomic or dynamic features in the reconstruction through a kernel matrix (15–17). A better alternative to the indirect method is direct parametric reconstruction, which estimates the tracer kinetic parameters directly from the dynamic PET sinograms because noise in PET sinograms can be accurately modeled. It has been shown that direct reconstruction methods can improve the image quality and signal-to-noise ratio of parametric images (18–20).
In this work, we aimed to demonstrate the first total-body parametric imaging on a human subject using the uEXPLORER scanner and the performance improvement using kernel reconstruction and direct reconstruction in comparison with indirect reconstruction. In this first study, we focused on the linear Patlak graphical model, which has been widely used to analyze the accumulation and retention of 18F-FDG-6-phosphate that occurs in proportion to the rate of glycolysis in specific tissues (21). For comparison, we studied the indirect Patlak method based on either ordered-subsets expectation maximization (OSEM) reconstruction or kernel reconstruction and also the direct Patlak reconstruction using the nested algorithm (22). The contrast-versus-noise curve was evaluated for the resulting parametric images to quantify the gain in noise reduction by kernel regularization and direct parametric reconstruction.
MATERIALS AND METHODS
Scanner Description
The uEXPLORER scanner has an AFOV of 194 cm and a transaxial field of view of 68.6 cm (9). It is constructed with 8 axial unit rings (each with an AFOV of 24 cm) formed from 24 detector modules with an inner ring diameter of 78.6 cm. Each module consists of 5 × 14 detector blocks with an array of 7 × 6 lutetium-yttrium-oxyorthosilicate scintillator crystals. Each crystal has dimensions of 2.76 × 2.76 × 18.1 mm. The uEXPLORER has time-of-flight (TOF) capability, with a system timing resolution of approximately 430 ps and energy resolution of 11.7% (7). A summary of the physical characteristics of the uEXPLORER is provided in Table 1. The maximum difference of ±4 unit rings was in use in the fully 3-dimensional data acquisition, with a maximum axial acceptance angle of 57°. A unit-difference–dependent coincidence time window was implemented (11), ranging from 4.5 ns (unit difference of 0) to 6.9 ns (unit difference of 4). The maximum ring difference of 4 unit rings was based on a previous simulation (23) showing that the noise-equivalent count rate peaks with a maximum acceptance angle of about 55°. Because of the attenuation effect of an adult human body, only 1% prompts from the body are rejected at this maximum acceptance angle.
Physical Characteristics of uEXPLORER Scanner
Quantitative Image Reconstruction and Correction
We developed quantitative image reconstruction software for the EXPLORER scanner (24). A TOF list-mode iterative reconstruction method was used to take advantage of the statistical and physical models of the PET imaging process. We modeled the system response by combining a calculated system matrix and a measured blurring kernel (25). The image-domain point-spread function was estimated by reconstructing simulated point sources at 11,532 transaxial and axial locations over the field of view. A component-based normalization was performed using a 2-m-long water cylinder phantom to estimate the crystal efficiencies and plane efficiency (26). The attenuation correction factors were obtained from a coregistered CT scan acquired using the CT component of the scanner. A Monte Carlo multiple-scatter simulation technique was implemented on a graphics processing unit to provide a scattered coincidence estimation (27). For random coincidence correction, the delayed coincidence method was used. Dead-time correction was not included in this initial study. Because of the huge number of lines of response in this scanner (>90 billion before accounting for TOF), TOF sinograms are impractical to store. Instead, a block-based TOF sinogram is used to reduce the storage size of scatter and random sinograms (24). For each unit pair, we grouped 1 transaxial block and 2 axial blocks to form 7 × 12 crystals as 1 element; the resulting compressed TOF sinogram has 77 radial bins, 60 angular bins, 7 × 7 block ring pairs, and 27 TOF bins (273 ps wide). We interpolated the scatter and random mean from the compressed TOF sinogram for the list-mode image reconstruction (24).
Evaluation Studies
Human Subject Study
We conducted the first human dynamic total-body PET study on the uEXPLORER. A healthy woman was recruited (61 y old; height, 156 cm; weight, 56 kg) and gave informed consent under the guidance of the Ethics Board of Zhongshan Hospital. A 1-h dynamic scan was performed immediately after an intravenous injection of 256 MBq of 18F-FDG via a vein near the ankle. Approximately 61 billion prompt coincidences and 41 billion delayed coincidences were recorded. The random fraction (RF) ranged from 63% to 70% during the acquisition time because of the changes in tracer distribution and radioactive decay (Fig. 1A). To exploit the high temporal resolution of this scanner, we divided the dataset into 187 temporal frames: 60 × 1 s, 30 × 2 s, 20 × 3 s, 12 × 10 s, 50 × 30 s, and 15 × 120 s.
(A) Count rate and random fraction of 1-h 18F-FDG dynamic scan of human subject. (B) Maximum-intensity projection of selected dynamic OSEM reconstructed images (1-s and 120-s frames). SUV images are shown in inverse gray scale with maximum set to 100 and 10. RF = random fraction.
Comparison Study
For quantitative image reconstruction, corrections (normalization, attenuation, randoms, scatter, and resolution model) were implemented in the forward model. Dynamic data were reconstructed using OSEM and kernel-based expectation maximization (kernel-EM). We used 3 composite frames (early 10 min, mid 20 min, and late 30 min) to extract image features for constructing the kernel matrix (15). Images were reconstructed into a 239 × 239 × 679 matrix with 2.85-mm cubic voxels. The linear Patlak graphical method was adopted to analyze the 18F-FDG kinetics in different tissues using the last 30 min of data (21). For comparison, indirect Patlak analysis from the reconstructed frames and direct reconstruction using the nested algorithm (22) were conducted. The input function was extracted from an ROI placed over the descending aorta instead of the left ventricle to minimize blurring effects due to cardiac motion and spillover from the myocardium. Because we did not know the ground truth for the value of Ki (net influx rate) in each voxel, we quantified the performance using a contrast-versus-noise curve in 2 different body regions. The first one was in the brain, where contrast was measured between the gray matter and the white matter and noise was calculated as the SD of the pixels within a uniform ROI in the white matter. The second one was in the thorax, where we measured the contrast of a focal region of uptake in the collarbone and the noise was calculated using the same white matter ROI. The high uptake in the collarbone was confirmed by a delayed scan acquired at 4 h after injection (9).
RESULTS
Dynamic Image Reconstruction
OSEM Reconstruction
The counting rate as a function of time from the 18F-FDG dynamic scan on the human subject is shown in Figure 1A. The total prompt rate (including trues, scatter, and randoms) peaked at 27.9 million counts per second at 6 s after injection as the radiotracer traveled to the cardiorespiratory system and then gradually dropped as the tracer distributed throughout the body, roughly following the decay of 18F. Figure 1B shows the anterior view of the maximum-intensity projection of OSEM reconstructed images of 6 selected temporal frames: 5 early 1-s frames from 17 to 21 s and the last 2-min frame from 58 to 60 min. These dynamic images show good image quality with low noise, even for the 1-s frames. In the high-temporal-resolution dynamic images, we can clearly see the tracer bolus entering the left ventricle first and then being pumped into the aorta. The temporal delay of the tracer bolus within the arterial tree is also evident.
Image-Derived Arterial Input Function (AIF)
We extracted the time–activity curves from 5 blood regions (left ventricle, aorta, carotid artery, brachial artery, and femoral artery) and 4 major organs/tissues (myocardium, liver, gray matter, and white matter). The results in Figure 2 show that the left ventricle is the earliest ROI to peak. The measured delay between the left ventricle and aorta is about 1–2 s, and the time delay between the aorta and brachial and femoral arteries is about 5 s. The arterial curves measured from the carotid, brachial and femoral arteries have lower peaks than the peak of the aorta because of dispersion and partial-volume effects. The time–activity curves are smooth in the ROIs of gray matter and white matter but not in the ROIs of myocardium and liver, because of cardiac motion and respiratory motion. On the basis of motion and partial-volume considerations, the ROI over the aorta was selected for all parametric analyses.
OSEM image–derived ROI-based blood input functions and time–activity curves (TACs). (Left) AIFs from different ROIs during first 2 min. (Middle) AIFs during whole 1-h scan zoomed in on 0–12 kBq/mL scale. (Right) Time–activity curves of major organs/tissues of interest.
Kernel-EM Reconstruction
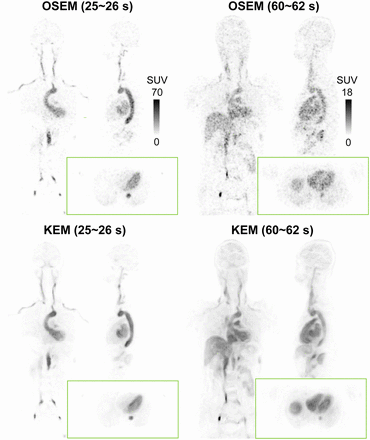
Figure 3 shows maximum-intensity projections of 3 composite images of early 10-min, mid 20-min, and late 30-min scans, which have low noise due to the large number of acquired counts (11.3, 20.2, and 28.6 billion prompts, respectively). These 3 composite images were applied to calculate the kernel matrix using a Gaussian function for the kernel-EM reconstruction (15). Figure 4 shows the comparison of orthogonal slices between OSEM and kernel-EM reconstructions at 2 selected early frames (25–26 s and 60–62 s). We can see that the image quality of these short temporal frames was greatly improved using the kernel method, with enhanced contrast between organs and reduced noise in uniform regions.
Reconstructed dynamic composite SUV images (OSEM maximum-intensity projections) of early 10-min, mid 20-min, late 30-min scans. Images were used to extract features to construct kernel matrix. SUV images are shown in inverse gray scale with maximum set to 9.
Coronal, sagittal, and transaxial slices of OSEM and kernel-EM (KEM) reconstructions of early 1-s frame (25–26 s) and 2-s frame (60–62 s).
Parametric Imaging
Indirect OSEM Versus Kernel-EM Versus Direct Reconstruction
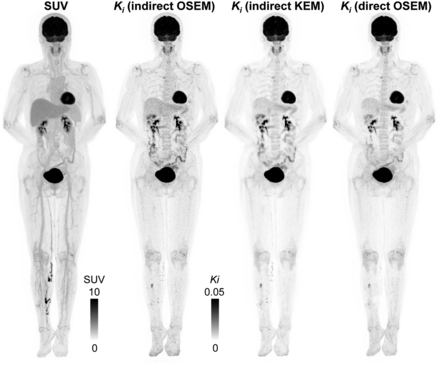
Total-body parametric images were obtained using the indirect parametric estimation based on either the OSEM or kernel-EM images and by direct parametric reconstruction. Figure 5A shows the orthogonal slices of the Patlak Ki (slope of the Patlak plot) images. Because of the increased sensitivity of uEXPLORER, even the OSEM indirect method can generate high-quality Patlak images. In comparison, the kernel-EM indirect Patlak and direct Patlak reconstructions further improve the parametric image quality, with a better contrast-versus-noise tradeoff, over the indirect method. There is excellent delineation of the smaller structures in cerebral, cardiac, and vertebral regions. The high-spatial-resolution images were reconstructed without incurring a high noise penalty. These observations are consistent with the quantitative results shown below.
(A) Coronal, sagittal, and transaxial slices of Patlak slope Ki reconstructed by indirect OSEM, indirect kernel-EM (KEM), and direct OSEM. (B) Ki contrast of gray matter to white matter vs. noise curves. SD was calculated in large ROI inside white matter. (C) Ki contrast of focal uptake lesion (red circles) in collarbone to white matter vs. noise curves. SD was calculated using same white matter ROI as in (B). Ki images are shown in inverse gray scale with maximum set to 0.06.
ROI Quantification
Figure 5B shows the Ki contrast between the gray matter and white matter versus the SD tradeoff. The indirect OSEM Patlak has the highest noise level at the same Ki contrast ratio. Both the indirect kernel-EM and direct OSEM Patlak can achieve high image contrast with reduced noise. The indirect kernel-EM also has a fast convergence speed with a much shorter reconstruction time than the direct OSEM Patlak. Figure 5C shows the contrast-versus-noise curve for the focal uptake in the collarbone. The relative performance between the different methods remains the same, with the kernel-EM and direct reconstruction achieving lower noise at the same contrast level than the indirect OSEM reconstruction.
SUV Versus Patlak Slope
Figure 6 shows maximum-intensity projections of the total-body SUV (late 30 min) and Patlak Ki images using different methods. We can see some correlation between SUV and Patlak Ki, but the Patlak Ki images have better tissue contrast without the interference of blood vessels. Furthermore, we also include a delayed scan of the same patient at 4 h after injection, which confirms the high focal uptake in the right sternoclavicular joint (Fig. 7). This result suggests that it may be possible to use parametric images to improve lesion conspicuity without the inconvenience of late time-point scanning.
Maximum-intensity projections using last 30 min of data. Indirect OSEM Patlak slope Ki uses 3 iterations and 10 subsets. Indirect kernel-EM (KEM) Patlak slope Ki uses 3 iterations and 10 subsets. Direct OSEM Patlak slope Ki uses 12 iterations and 10 subsets, with 10 subiterations of parametric update.
Total-body dynamic 18F-FDG PET and parametric imaging. (A) Static PET images and fused PET/CT image to confirm high uptake at right sternoclavicular joint, degenerative in nature. Both PET images were obtained with 14-min duration. (B) Maximum-intensity projections of OSEM SUV image and indirect Patlak slope Ki image. Latter image is from scan 30–60 min after injection. Red circles indicate high 18F-FDG uptake in right sternoclavicular joint. p.i. = after injection.
DISCUSSION
Parametric imaging from dynamic PET has the benefit of improving the accuracy of tracer kinetic assays of biologic and pharmacologic processes, which can only be inferred indirectly from static uptake PET images. In this work, we demonstrated total-body parametric imaging on the uEXPLORER scanner.
A major advantage of using the uEXPLORER is the simultaneous total-body coverage. Dynamic PET imaging using existing whole-body scanners is often limited to one bed position, which is incompatible with the standard whole-body imaging protocol. Although the multiple-bed-position, multipass approach can provide dynamic imaging with an extended AFOV, its temporal resolution is limited by bed motion and the sensitivity of the scanner and it cannot capture fast tracer dynamics (e.g., total-body perfusion with a highly diffusible tracer). Accurately tracking the time information of the bed motion is also required to obtain consistent results for parametric images (4). In contrast, the uEXPLORER scanner allows dynamic imaging of the entire body with high temporal resolution because there is no bed motion and the system has high sensitivity to yield a high signal-to-noise ratio in short time frames. With its total-body coverage, the uEXPLORER scanner simplifies the protocol for dynamic PET, and every scan can be performed in a dynamic fashion without any sacrifice in the axial coverage. The total-body coverage also allows the image-derived AIF to be determined from the aorta regardless of the organ of interest, which is impossible to determine on existing whole-body PET scanners except for regions near the heart. Although it was expected that the AIF would be dependent on the sampling position, simultaneously measuring AIF at various sampling sites has not been possible because of the limited axial coverage of current clinical whole-body PET scanners. Using the uEXPLORER, we measured AIFs simultaneously from different sites inside the human body. The results show differences between the AIFs. Considering motion and partial-volume effects, we chose the AIF from the aorta for all parametric analysis. Finding a way to use the AIFs from different arteries synergistically for total-body parametric imaging is an ongoing area of research.
Another major advantage of total-body dynamic imaging is the high sensitivity. Either with the step-and-shoot mode or the continuous-bed-motion mode, the whole-body multiple-bed-position, multipass approach loses sensitivity for photon detection and misses spatiotemporal information between passes. Total-body dynamic PET can significantly improve the counting statistics because all organs are inside the field of view during the entire scan. The combination of total-body coverage and high sensitivity allows dynamic imaging with much higher temporal resolution than is possible on existing PET scanners. High temporal resolution will be especially beneficial when nonlinear or more complex kinetic models are used.
There are a few limitations in this current work. First, we focused on the Patlak graphical plot in our initial total-body parametric analysis because of its simplicity. This linear model is limited to irreversible tracer uptake, which can represent the biologic behavior of only certain organs (tissues and cells) and specific tracers. In many cases, we need to investigate nonlinear kinetic models. This is an ongoing and future effort. Second, our preliminary human study recruited only a healthy volunteer. It is necessary to conduct patient studies in the future to evaluate the benefits and potential of parametric imaging in oncologic studies. Third, because of the long scan duration, subject motion was observed in the dynamic images. We are currently developing motion correction methods either as a postprocessing technique or in a motion-compensated reconstruction framework.
CONCLUSION
We have successfully demonstrated total-body parametric imaging on a human subject using the uEXPLORER scanner. In doing so, we evaluated the benefits of kernel reconstruction and direct reconstruction methods to the quality of parametric images. With its total-body coverage and high sensitivity, the uEXPLORER scanner allows us to realize the full quantitative potential of PET imaging.
DISCLOSURE
Ramsey Badawi and Simon Cherry have a research agreement funded by United Imaging Healthcare. UC Davis also has a uEXPLORER sales-based revenue-sharing agreement with United Imaging Healthcare. This work was supported in part by the National Institutes of Health under grant R01 CA206187 and UC Davis Innovative Development Award (IDA). No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: Is the uEXPLORER capable of total-body parametric imaging, and can image quality and kinetic parameter estimation be improved by direct and kernel reconstruction of the uEXPLORER data?
PERTINENT FINDINGS: The reconstructed total-body parametric images show high image quality. The image quality can be further improved using kernel-based regularization and direct parametric image reconstruction.
IMPLICATIONS FOR PATIENT CARE: With its total-body coverage and high sensitivity, the uEXPLORER allows us to realize the full quantitative potential of PET imaging.
Acknowledgments
We acknowledge the contributions of all team members from UC Davis, United Imaging Healthcare, and Zhongshan Hospital. We thank Drs. Terry Jones and Michael Phelps for their helpful comments on the manuscript. We thank Dr. Lorenzo Nardo for confirming the high-uptake region in the SUV images.
Footnotes
Published online Jul. 13, 2019.
- © 2020 by the Society of Nuclear Medicine and Molecular Imaging.
REFERENCES
- Received for publication May 1, 2019.
- Accepted for publication July 6, 2019.