Abstract
251
Background: Individuals with Down syndrome (DS) are at increased risk of developing Alzheimer’s disease (AD) and show the earliest signs of amyloid-β (Aβ) deposition in the striatum. The metric for tracking amyloid burden using PET radiotracers frequently uses average SUVr in signature regions of the brain specific to Aβ deposition. An alternative index of amyloid load (AβL) was recently developed (Whittington 2018) as a template-based approach of quantifying Aβ burden from static PET data with high sensitivity to detect Aβ change. The algorithm determines AβL and a non-specific binding coefficient (ns) given inputs of SUVr and canonical images of Aβ carrying capacity (K) and tracer non-specific binding (NS). Objective: The purpose of this study was to implement the AβL algorithm in the DS population using DS-specific [11C]PiB PET templates for spatial normalization, K and NS with the inclusion of striatal PiB binding in the model parameters. Additionally, a cutoff for PiB(+) based on AβL were determined from the dataset.
Methods: Participants with DS (N=100; 38.5±8.2 yrs) were enrolled as non-demented, with some converting to MCI and AD during the course of the study. All participants underwent a 50-70 minute [11C]PiB scan in addition to T1w-MRI. A subset of participants (n=52, 35, and 7) underwent two, three, and four PiB scans, respectively (2.3±0.6 years apart). ROI definition was performed through segmentation of the T1w-MRI using FreeSurfer v5.3.0. SUVr images were generated from the PET data using cerebellar gray matter as the region for normalization. Global PiB was computed as the average SUVr from the striatum and cortex. A sigmoidal growth curve (fit by the logistic growth model of Aβ accumulation) describing AD chronology (30 year scale) was generated by integrating longitudinal rates of striatal PiB change (Figure 1). The PET data were spatially normalized to MNI152 space using a DS-specific PiB PET template, smoothed with a 4 mm Gaussian kernel, and a time point in AD chronology was determined for each participant. Parametric images of K and NS were generated by applying the logistic growth model at the voxel level to the SUVr data. AβL and ns were calculated using the AβL algorithm for each participant. Thresholds for PiB(+) were determined from sparse k-means clustering with resampling as implemented on SUVr data.
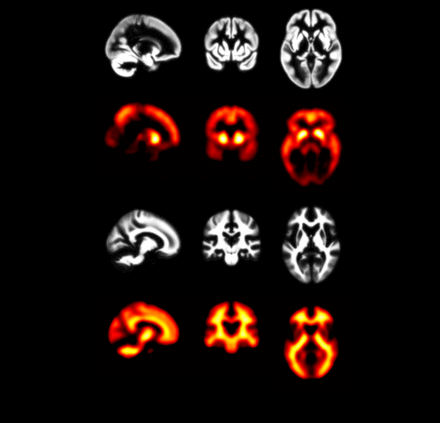
Results: DS-specific maps for K and NS (Figure 2) were consistent with the known Aβ pathology (with highest voxel intensities in the striatum) and white matter PiB binding. A strong positive correlation was observed between AβL and global PiB SUVr (Pearson’s r = 0.97) and no correlation was observed between ns and SUVr (r = -0.09; Figure 3). An AβL cutoff for PiB(+) of 23.9% was determined from k-means clustering. Conclusion: These results highlight the promise of AβL for quantification of Aβ burden in DS. Unique to this population (compared to sporadic AD) is the striatum displaying the highest carrying capacity. Analysis is ongoing to evaluate longitudinal change in AβL in this cohort to better characterize the natural history of Aβ in DS.