Abstract
244
Introduction: Coronary plaque 18F-Sodium Fluoride (18F-NaF) images need to be acquired over long time (up to 30 minutes) to obtain diagnostic image quality due to low signal and noise in the images but this is inconvenient to patient, decreases patient throughput and can result in patient motion. In this study we investigate the feasibility of reconstructing images equivalent to 30 min 18F-NaF-PET acquisitions using 1/10th of the acquired counts using deep learning (DL) computer techniques.
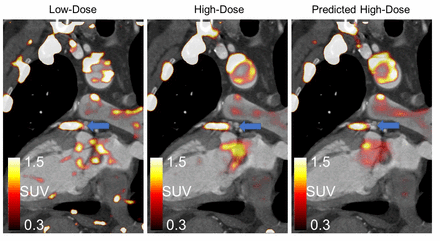
Methods: To train the DL model, we included 132 patients recruited for 30 min long 18F-NaF coronary plaque 250 MBq PET imaging in a hybrid PET/CT Angiography (CTA) protocol. Two image sets were reconstructed for each scan, a static image using all the data (Full-dose) and an image set using 10% of the acquired PET counts (Low-dose) using list mode data. The DL model (U-net) was trained to predict the Full-dose scan from the Low-dose scans slice by slice (creating third image dataset - Predicted High Dose), using 10-fold cross validation for parameter optimization. Lesion uptake was quantified from spherical Volume of Interest (VOI) (radius=5mm), while background activities were obtained in the right atrium using a cylindrical VOI (length=15mm, radius=10mm). Lesions with target-to-background ratios (TBR) ≥1.25 were considered 18F-NaF-active, while TBR<1.25 were considered 18F-NaF-negative. All lesions were classified from the Full-dose data sets. A separate test group consisting of 20 patients with known coronary artery disease were analyzed in this study with the DL reconstruction. We report the number of coronary lesions as well as the signal-to-noise ratios (SNR) obtained for the three datasets (Full-dose, Low-dose and Predicted-High-Dose).
Results: A total of 55 coronary lesions (25 18F-NaF-positive) were identified on the 20 Full dose scans. The Low-dose datasets had significantly lower SNR (8.9±4.8), in the lesions compared to Full-dose data (19.5±12.8), p<0.001, while no difference were observed for the Predicted-High-Dose SNR (16.1±12.1, p=0.14). Figure shows examples of High-Dose, Low-Dose and Predicted-High-Dose reconstructions.
Conclusions: Using DL, it is possible to reconstruct images from 3-minute scan data with SNR equivalent to 30 min long acquisitions. This technique may help reducing the long acquisition times in current coronary PET clinical studies.