Abstract
Previous studies have demonstrated the feasibility of absolute quantification of dynamic 123I-metaiodobenzylguanidine (123I-MIBG) SPECT imaging in humans. This work reports a simplified quantification method for dynamic 123I-MIBG SPECT using practical protocols with shortened acquisition time and voxel-by-voxel parametric imaging. Methods: Twelve healthy human volunteers underwent five 15-min dynamic SPECT scans at 0, 15, 90, 120, and 180 min after 123I-MIBG injection. List-mode SPECT data were binned into 29 frames and reconstructed with corrections for attenuation, scatter, and decay. Population-based blood-to-plasma correction and metabolite correction were applied to the image-derived input function. Likelihood estimation in graphical analysis (LEGA) was used as a simplified model to obtain volume of distribution (VT) values, which were compared with those obtained with the reversible 2-tissue (2T) compartment model. Three simplified protocols were evaluated with 2T and LEGA using a 30-min scan started simultaneously with tracer injection plus a 15-min scan at 90, 120, or 180 min after injection. Voxel-by-voxel LEGA fitting was applied to the aligned dynamic images using both the full protocol (five 15-min scans) and the simplified protocols. Results: Correlation analysis (y = 0.955x + 0.547, R2 = 0.997) and Bland–Altman plot (mean difference, −0.8 mL/cm3; 95% limits of agreement, [−2.5, 1.0] mL/cm3; normal VT range, 29.0 ± 12.4 mL/cm3) showed that LEGA can be used as a simplified model of 2T for 123I-MIBG. High-quality VT parametric images could be obtained with LEGA. Region-of-interest (ROI) modeling and parametric imaging results were in excellent agreement as determined by correlation analysis (y = 0.999x − 1.026, R2 = 0.982) and Bland–Altman plot (mean difference, −1.0 mL/cm3; 95% limits of agreement, [−4.2, 2.1] mL/cm3). VT correlated reasonably well between all simplified protocols and the full protocol with LEGA but not with 2T. The VT results were more reliable when there was a longer interval between the 2 acquisitions in the simplified protocols. Conclusion: For ROI-based kinetic modeling and parametric imaging, reliable quantification of dynamic 123I-MIBG SPECT can be achieved with LEGA using a simplified protocol of a 30-min scan starting with tracer injection plus a 15-min scan no earlier than 180 min after injection.
Myocardial sympathetic imaging with 123I-metaiodobenzylguanidine (123I-MIBG) has been extensively used for assessing patients with congestive heart failure, ischemic heart disease, and primary arrhythmic disease (1–4). The heart-to-mediastinum ratio calculated from 2-dimensional (2D) planar imaging has remained the conventional standard for 123I-MIBG quantification. However, heart-to-mediastinum ratios obtained from 2D planar imaging are only semiquantitative because of the inclusion of background activity in the quantification of the activity within the heart from a 2D region of interest (ROI) and the lack of corrections for attenuation and scatter. The use of 3-dimensional (3D) SPECT imaging for 123I-MIBG quantification has been applied more recently to overcome these limitations and make regional analysis of myocardial uptake feasible (5–11). However, myocardial 123I-MIBG uptake changes with time, especially in heart failure patients, with the myocardial activity decreasing continuously even after 3 h after injection (12,13). The use of 4-dimensional (4D) dynamic SPECT imaging with tracer kinetic modeling can provide intrinsic physiologic parameters for quantifying sympathetic nerve activity, which is independent of the imaging time points and has the potential to be more sensitive and accurate than the estimation of a 2D or 3D heart-to-mediastinum ratio. This dynamic 4D approach can facilitate standardization of quantification of 23I-MIBG images for wider clinical applications.
Our group recently reported the feasibility of absolute quantification of sympathetic activity with 4D dynamic 123I-MIBG SPECT imaging using a stationary dedicated cardiac cadmium zinc telluride SPECT/CT scanner (Discovery NM/CT 570c; GE Healthcare) over a 3- to 4-h period (13). Population-based blood-to-plasma ratio correction and metabolite correction techniques were proposed and demonstrated to be feasible for tracer kinetic modeling in a healthy human population. The volume of distribution (VT) values (range, 15.0–53.8 mL/cm3; mean, 29.0 ± 12.4 mL/cm3) for 123I-MIBG in myocardium were reported for 12 healthy humans using a reversible 2-tissue (2T) compartment model.
However, 3–4 h of dynamic imaging may not be clinically feasible. To establish a more clinically practical quantitative approach, we have extended our previously developed dynamic 123I-MIBG technique (13) and investigated simplified acquisition protocols using only 2 scans and a shorter total acquisition time with a simplified model. We have investigated, for what is to our knowledge the first time, a voxel-by-voxel parametric imaging technique for analysis of 123I-MIBG SPECT studies that will enable future regional analysis of clinical VT images, potentially providing additional information on the heterogeneity of myocardial sympathetic activity. This study used the same dataset from 12 healthy humans as was used in a previous report (13).
MATERIALS AND METHODS
Study Subjects and Dynamic SPECT Image Generation
Twelve healthy humans (7 male and 5 female; mean age ± SD, 48 ± 13 y) received an intravenous bolus injection of 233 ± 67 MBq of 123I-MIBG (AdreView; GE Healthcare). Five 15-min dynamic SPECT scans were acquired on the Discovery NM/CT 570c scanner starting at 0, 15, 90, 120, and 180 min after injection. The subjects remained in the scanner for the first 2 scans and then left the scanner between the subsequent scans to take breaks. An unenhanced CT scan was acquired for attenuation correction at the end of the last SPECT acquisition. The study was approved by the institutional review board, and all subjects signed an informed consent form. The list-mode SPECT data from all 5 scans were binned offline into 29 frames (15 × 20 s and 14 × 300 s). The dynamic images were reconstructed using maximum-likelihood expectation maximization (80 iterations) with corrections for attenuation, scatter, and decay. Detailed information about the study subjects and the image reconstruction process was previously reported (13).
Kinetic Modeling
For each subject, an image-derived input function and myocardial time–activity curve were obtained by drawing 3D ROIs on the left ventricle blood pool cavity and on the left ventricle myocardium, respectively. To reduce the effect of body motion between scans, the ROIs of each scan were generated separately with the same volumes. Population-based blood-to-plasma ratio correction and metabolite correction, which were previously found feasible for a healthy human population (13), were applied to the image-derived input function for tracer kinetic modeling. 2T was used, which has been previously identified as the optimal model for 123I-MIBG modeling (12,13). However, the implementation of 2T is computationally intensive, particularly for the voxel-by-voxel fitting required to generate parametric images. 2T is also sensitive to the noise in the dynamic images. A simplified model is often applied to address these issues. The commonly used Logan graphical analysis (14) can estimate VT more rapidly and robustly than 2T but suffers from noise-induced bias (15,16). To overcome this limitation, we used likelihood estimation in graphical analysis (LEGA) (17,18), as previously implemented (19), which can robustly estimate VT by accounting for the noise in the original data domain.
The estimated myocardial time–activity curve using LEGA,  , can be described as follows:
, can be described as follows: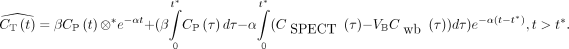 Eq. 1β and α are 2 intermediate parameters, defined by α = −1/b, where b is the y-intercept of the Logan plot and
Eq. 1β and α are 2 intermediate parameters, defined by α = −1/b, where b is the y-intercept of the Logan plot and  .
.  is the time after which the LEGA equation holds (i.e., when the Logan plot is considered linear).
is the time after which the LEGA equation holds (i.e., when the Logan plot is considered linear).  is a customized convolution operator using only data after
is a customized convolution operator using only data after  (i.e.,
(i.e.,  ).
).  is the plasma time–activity curve after metabolite correction used as the input function.
is the plasma time–activity curve after metabolite correction used as the input function.  is the whole-blood time–activity curve.
is the whole-blood time–activity curve.  is the blood volume term accounting for the spill-in effect from the blood pool to the myocardium.
is the blood volume term accounting for the spill-in effect from the blood pool to the myocardium.  is the myocardial time–activity curve obtained from the SPECT images, including the blood volume spill-in.
is the myocardial time–activity curve obtained from the SPECT images, including the blood volume spill-in. 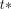 was selected to be 7.5 min through a pilot investigation using multiple values applied on the data for 12 subjects, and
was selected to be 7.5 min through a pilot investigation using multiple values applied on the data for 12 subjects, and  was set to be the mean value 0.3 mL/cm3 obtained from 2T (13).
was set to be the mean value 0.3 mL/cm3 obtained from 2T (13).
The fitted parameters  and α were estimated using weighted least squares with a Marquardt–Levenberg algorithm (20) as follows:
and α were estimated using weighted least squares with a Marquardt–Levenberg algorithm (20) as follows: Eq. 2
Eq. 2 Eq. 3
Eq. 3 is the estimation of
is the estimation of  .
.  and
and  are the start time and end time of frame i. k is the start frame number (i.e.,
are the start time and end time of frame i. k is the start frame number (i.e.,  ), and n is the end frame number (i.e., the total number of frames).
), and n is the end frame number (i.e., the total number of frames).  is the weighting factor of frame i calculated by:
is the weighting factor of frame i calculated by: Eq. 4where
Eq. 4where  is the total projection count number of frame i and
is the total projection count number of frame i and  is the decay correction factor of frame i.
is the decay correction factor of frame i.
Simplified Acquisition Protocols
In clinical practice, it is often difficult to implement dynamic 123I-MIBG imaging that incorporates five 15-min scans over 3–4 h. To reduce the number of scans and the total acquisition time, we investigated 3 different simplified acquisition protocols that make use of fewer scans (2 scans) and a shorter total acquisition time (45 min). Accordingly, a 30-min scan starting immediately after 123I-MIBG injection (including two 15-min scans starting at 0 and 15 min) was incorporated with a 15-min late-phase scan at 180 min (30 + 180 min), 120 min (30 + 120 min), or 90 min (30 + 90 min) after injection.
For the full protocol with all five 15-min scans, the SPECT data were binned into 29 frames (15 × 20 s and 14 × 300 s). For the simplified acquisition protocols, the data of the first 30-min scan were binned into 20 frames (15 × 20 s and 5 × 300 s) and those of the late-phase 15-min scan were binned into 3 frames (3 × 300 s). We investigated these 3 simplified protocols using both 2T and LEGA.
Parametric Imaging
To establish methods to measure regional VT to evaluate the heterogeneity of 123I-MIBG uptake in the myocardium, parametric imaging was implemented with voxel-by-voxel modeling on the dynamic SPECT images. For parametric imaging, only LEGA was used in this work, because 2T is time-consuming and sensitive to noise on the voxel level. Images were registered before the voxel-by-voxel fitting, because the subject left the scanner between scans. To minimize the registration error due to the high noise and the truncation artifact in the SPECT image, we implemented the registration based on the CT-derived attenuation maps instead of SPECT images. First, the attenuation map was registered to each SPECT image (without attenuation correction) on a GE Healthcare Xeleris workstation using the attenuation correction quality control protocol. The 15-min SPECT scan starting at 15 min after injection was chosen as the reference scan. The attenuation map of each scan was rigidly registered to that of the reference scan using BioImage Suite software with the normalized mutual information as the similarity metric (21). Then, the transformation matrix derived from attenuation map registration was applied to the SPECT images of each scan to register all the dynamic SPECT images to the reference scan. Voxel-by-voxel fitting was implemented on the aligned dynamic SPECT images using LEGA. The input function used for parametric imaging was the same as that used in the ROI-based modeling process. The simplified protocols were also investigated in the parametric imaging study.
Statistical Methods
Correlation values and least-squares fits were calculated for the 12 healthy humans, first, between ROI VT with 2T and with LEGA; second, between ROI VT and mean myocardial VT using parametric imaging where both were derived using LEGA; and third, between ROI VT with the simplified protocols (with both 2T and LEGA) and the full protocol with 2T (denoted as the gold standard). Bland–Altman analysis (22) was also used to further investigate the agreement of the methods.
On parametric images, the normalized root-mean-squared error (NRMSE) of the difference in myocardial VT between the simplified and full protocols was calculated as Eq. 5where
Eq. 5where  and
and 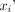 are the VT of voxel i in the myocardial ROI of the parametric images obtained with the full protocol and with the simplified protocol, respectively, and N is the total number of voxels in the myocardial ROI.
are the VT of voxel i in the myocardial ROI of the parametric images obtained with the full protocol and with the simplified protocol, respectively, and N is the total number of voxels in the myocardial ROI.
RESULTS
ROI-Based Kinetic Modeling with Simplified Model
Figure 1 shows typical fitting results with 2T and LEGA for a sample subject. The fits of the myocardial time–activity curves nearly overlapped. The ROI-based VT obtained using 2T and LEGA are compared in Figure 2. Figure 2A shows that the obtained VT was almost identical in each of the 2 models. An excellent linear relationship was obtained (y = 0.955x + 0.547, R2 = 0.997), indicating that LEGA can be used as a simplified model of 2T for 123I-MIBG modeling. The Bland–Altman plot shown in Figure 2B further confirmed this finding. The mean VT difference between 2T and LEGA was −0.8 mL/cm3, and the 95% limits of agreement were [−2.5, 1.0] mL/cm3, which are small compared with the VT range of this population (15.0–53.8 mL/cm3; mean, 29.0 ± 12.4 mL/cm3). Although a negative slope (−0.044) significantly different from zero (P = 0.04) was observed in the Bland–Altman plot, the range of VT difference (≤2.5 mL/cm3) was small when considering the VT range (≤53.8 mL/cm3).
Typical time–activity curve (TAC)–fitting results for 2T and LEGA ( = 7.5 min) in healthy human for total of 3 h (A) and after 7 min (B).
= 7.5 min) in healthy human for total of 3 h (A) and after 7 min (B).
Comparison of ROI-based VT obtained using 2T and LEGA: correlation result (A) and Bland–Altman plot (B).
ROI-Based Kinetic Modeling with Simplified Protocol
The ROI-based results using 3 simplified protocols for 2T and LEGA are shown in Figure 3 and were compared with the gold standard VT, obtained with the full protocol with 5 scans and 2T. By using 2T, there was no apparent correlation of VT between the simplified protocols and the full protocol. In contrast, strong correlations of VT could be obtained using LEGA, indicating that LEGA is more robust than 2T for the simplified protocols when analyzing less dynamic data with large gaps in between. The results showed that the protocol using only 2 scans with a shorter total acquisition time (45 min) is feasible for dynamic 123I-MIBG imaging using LEGA. Among these 3 simplified protocols, the VT results were more reliable when there was a longer interval between the 2 scans, because the R2 values derived from the “30 + 180 min” (0.931) and “30 + 120 min” (0.937) protocols appeared to be higher than the R2 value derived from the “30 + 90 min” protocol (0.814). However, no significant difference in R values (square root of R2) was found between the “30 + 180 min” and “30 + 120 min” protocols (P = 0.9) or between the “30 + 180 min” and “30 + 90 min” protocols (P = 0.1) using the Williams’ 2-tailed test (23). The slope of the “30 + 180 min” protocol was closer to 1 (0.885) than the slope of the “30 + 120 min” (0.489) or “30 + 90 min” (0.640) protocol, with a significant difference, since the linear regression slope between VT difference of the 2 simplified protocols and the gold standard VT was significantly different from zero (P = 0.0008 between “30 + 180 min” and “30 + 120 min”; P = 0.03 between “30 + 180 min” and “30 + 90 min”). Figure 4 shows the Bland–Altman plots between the simplified protocols with LEGA and the gold standard, which confirmed that the mean VT difference (−1.7 mL/cm3) was the smallest using the “30 + 180 min” protocol. In the Bland–Altman plots, the slopes (−0.67 and −0.36) of the differences were significantly different from zero (P = 3 × 10−6 and 0.03) for the “30 + 120 min” and “30 + 90 min” protocols, respectively, but such was not the case for the “30 + 180 min” protocol (slope, −0.088; P = 0.3). These results indicate that when LEGA is used as a simplified model, the “30 + 180 min” protocol can be selected as an optimized simplified protocol for a dynamic 123I-MIBG study in clinical practice.
Correlation results for ROI-based VT between gold standard (VT for full protocol with 2T) and simplified protocols (VT for protocols of 30 + 180 min, 30 + 120 min, and 30 + 90 min) with both 2T and LEGA.
Bland–Altman plots of ROI-based VT between gold standard (VT for full protocol with 2T) and simplified protocols (VT for protocols of 30 + 180 min, 30 + 120 min, and 30 + 90 min) with LEGA.
Parametric Imaging
Sample VT image of a healthy human with LEGA using the full protocol is shown in Figure 5 and compared with the 5-min static SPECT image at 3 h after injection. The results show that the LEGA model can generate VT parametric images with noise levels that are similar to those of static SPECT images, though more counts are included to generate parametric images.
Sample 5-min static SPECT image at 3 h after tracer injection and sample VT images with LEGA using different protocols for healthy human, shown in transverse, coronal, and sagittal planes. All VT images were scaled to same maximum.
Figure 6 shows the comparison results between the LEGA-derived VT using ROI-based modeling and the mean myocardial VT using parametric imaging for the 12 subjects. Figure 6A shows a strong linear correlation (y = 0.999x − 1.026, R2 = 0.982) between the ROI-based and parametric imaging results, demonstrating that parametric imaging of 123I-MIBG is feasible and accurate. The Bland–Altman plot in Figure 6B shows that the mean VT difference between ROI-based and voxel-based analysis was −1.0 mL/cm3, and the 95% limits of agreement were [−4.2, 2.1] mL/cm3, demonstrating consistent VT quantification. The slope (0.0086) of the differences in the Bland–Altman plot was not significantly different from zero (P = 0.8).
Comparison of VT obtained with LEGA using ROI-based modeling and myocardial VT obtained with parametric imaging for 12 subjects: correlation result (A) and Bland–Altman plot (B).
Parametric Imaging with Simplified Acquisition Protocol
Sample LEGA VT images of a healthy human with the simplified protocols are shown in Figure 5 and compared with the VT image with the full protocol. The VT image with the “30 + 180 min” protocol was nearly identical to that obtained with the full protocol, whereas the noise level in the VT images of the other 2 simplified protocols were much higher.
Figure 7 shows the normalized root-mean-squared errors of myocardial VT between the simplified protocols and the full protocol in parametric images with LEGA calculated by Equation 5. The mean normalized root-mean-squared error (±SD) was 0.24 ± 0.06 for the “30 + 180 min” protocol, 0.36 ± 0.11 for the “30 + 120 min” protocol, and 0.41 ± 0.11 for the “30 + 90 min” protocol. These results indicated a similar conclusion to that made for the ROI-based modeling, where the “30 + 180 min” protocol can be used as an optimized simplified protocol for clinical 123I-MIBG parametric imaging using voxel-by-voxel LEGA fitting.
Normalized root-mean-squared error of myocardial VT between simplified protocols and full protocol in parametric images with LEGA for each subject.
DISCUSSION
Our study investigated quantitative analysis of 123I-MIBG dynamic SPECT images using a simplified model (LEGA) for a healthy human population and compared this simplified model with 2T, which was identified as the optimal compartment model in our previous study. There was excellent agreement between the VT obtained from each of the 2 models, indicating LEGA can be used as a simplified model instead of 2T in clinical practice. The advantage of using LEGA is that the computation is much faster and more robust to noise than 2T, making LEGA more suitable for implementation in clinical practice. The successful use of a simplified model for analysis of dynamic 123I-MIBG SPECT images makes such a model possible for time-consuming and noise-sensitive parametric imaging using voxel-by-voxel fitting. In this study, the parametric image of VT obtained using LEGA was shown to be of high quality and may provide the possibility for regional analysis of myocardial 123I-MIBG uptake in future clinical investigations of the heterogeneity of myocardial sympathetic activation.
To implement this 123I-MIBG dynamic SPECT imaging technique in clinical practice, simplified acquisition protocols with fewer scans and a shorter total acquisition time were investigated. Three different simplified protocols were investigated using both ROI-based 2T and LEGA analysis. The results indicated that all 3 simplified protocols with 2T cannot provide reliable VT. However, the implementation of the simplified protocols was feasible using LEGA. We found that the protocol using an early 30-min acquisition and a delayed 15-min acquisition at 180 min after injection provided the VT estimate that best agreed with the gold standard VT using the full protocol. This finding may indicate that VT results are more reliable when there is a longer interval between the 2 acquisitions to capture the tracer kinetics. The simulation results reported in the supplemental data (supplemental materials are available at http://jnm.snmjournals.org) show that the VT results obtained with a delayed scan acquired at 3, 4, or 5 h were quite similar, indicating that the VT quantification is less sensitive to the exact acquisition time of the delayed scan, as long as it is acquired no earlier than 180 min after injection. This flexibility makes the proposed simplified protocol easy to implement clinically, because the delayed scan can be acquired whenever the SPECT scanner is available rather than at exactly 4 h after injection as in the current 2D heart-to-mediastinum ratio guideline. The VT images (Fig. 5) indicated that the simplified “30 + 180 min” protocol with voxel-by-voxel LEGA analysis provides a reliable parametric map of VT for clinical use.
In this study, direct registration between dynamic SPECT images did not provide satisfactory results, because of the noise and truncation artifact in the SPECT images during the parametric imaging process. Alternatively, an attenuation-map–based rigid registration method was implemented to align the dynamic SPECT images to compensate for body motion between scans. However, the body motion is likely to be nonrigid, because the subject left the scanner between scans, though the motion of the heart due to body movement is largely rigid. Our future work will focus on improving the registration accuracy by investigating the nonrigid body motion correction directly using the SPECT data. Respiratory and cardiac motion correction was not used in this study but, if implemented, might further improve the quantification accuracy of the VT image. Several innovative respiratory and cardiac motion correction methods have been proposed recently (24–28), and we will investigate adding a motion correction technique to our future study protocol.
The simplified quantification technique for analysis of dynamic 123I-MIBG SPECT images was validated using a healthy human population in this study. We have also applied the proposed technique to 4 heart failure patients. The results reported in the supplemental data indicate that simplified quantification of dynamic 123I-MIBG SPECT is also feasible in these patients. The parametric imaging technique is important because it allows for regional analysis of the patient images and thus may provide a more accurate measurement of VT when the kinetics differ between the normal and the defect zones of the myocardium. The limitation of this study was the small sample size. A prospective larger trial is needed with more healthy humans and heart failure patients.
CONCLUSION
Robust simplified quantification of dynamic 123I-MIBG SPECT images is feasible with LEGA. A simplified acquisition protocol comprising a 30-min dynamic scan that starts with the 123I-MIBG injection and a 15-min scan at 180 min after injection is robust and clinically practical for both ROI-based kinetic modeling and parametric imaging. High-quality parametric VT images can be obtained for healthy humans with the proposed simplified quantification technique, which might enable regional analysis of 123I-MIBG uptake directly from the VT image in clinical practice.
DISCLOSURE
This study was supported by American Heart Association Grant-in-Aid awards 13GRNT17090037 and 14GRNT19040010, and research contracts from GE Healthcare. No other potential conflict of interest relevant to this article was reported.
Footnotes
Published online Feb. 23, 2018.
- © 2018 by the Society of Nuclear Medicine and Molecular Imaging.
REFERENCES
- Received for publication September 14, 2017.
- Accepted for publication February 3, 2018.














