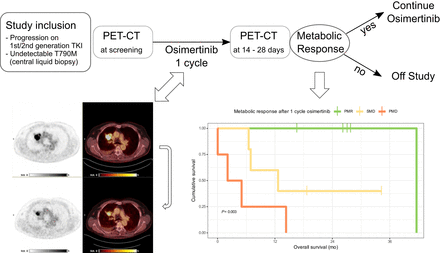
Visual Abstract
Abstract
Targeted therapy with epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) has established the precision oncology paradigm in lung cancer. Most patients with EGFR-mutated lung cancer respond but eventually acquire resistance. Methods: Patients exhibiting the EGFR p.T790M resistance biomarker benefit from sequenced targeted therapy with osimertinib. We hypothesized that metabolic response as detected by 18F-FDG PET after short-course osimertinib identifies additional patients susceptible to sequenced therapy. Results: Fourteen patients with EGFR-mutated lung cancer and resistance to first- or second-generation EGFR TKI testing negatively for EGFR p.T790M were enrolled in a phase II study. Five patients (36%) achieved a metabolic 18F-FDG PET response and continued osimertinib. In those, the median duration of treatment was not reached (95% CI, 24 mo to not estimable), median progression-free survival was 18.7 mo (95% CI, 14.6 mo to not estimable), and median overall survival was 41.5 mo. Conclusion: Connecting theranostic osimertinib treatment with early metabolic response assessment by PET enables early identification of patients with unknown mechanisms of TKI resistance who derive dramatic clinical benefit from sequenced osimertinib. This defines a novel paradigm for personalization of targeted therapies in patients with lung cancer dependent on a tractable driver oncogene.
The term personalized cancer therapy describes the administration of targeted cancer medicines on the basis of predictive biomarkers. Among solid tumors, non–small cell lung cancer (NSCLC) features the most diverse spectrum of globally approved precision medicines. Although the first line of personalized NSCLC therapies has a high likelihood of response, acquired resistance eventually limits their long-term benefit in most patients. Activating somatic epidermal growth factor receptor (EGFR) mutations define a lung cancer entity with high susceptibility to EGFR tyrosine kinase inhibitor (TKI) treatment. They are detected in 10%–15% of lung adenocarcinomas (>40% in East Asian populations), with predominance in nonsmokers and women. Analyses of tissue biopsy samples and circulating tumor DNA taken at progression have identified resistance mechanisms to NSCLC precision medicines (1–3). These may inform sequenced targeted therapies using non–cross-resistant agents (4) or drug combinations (5). Osimertinib treatment in EGFR p.T790M-associated acquired resistance is a prime example of sequenced personalized therapy (6). EGFR p.T790M is detected in approximately 60% of EGFR-mutant NSCLC progressing under first- (gefitinib, erlotinib) or second-generation (afatinib, dacomitinib) EGFR TKI (7). In those patients, sequenced osimertinib is superior to platinum-based chemotherapy (8). Sequenced osimertinib is not available to patients with undetectable EGFR p.T790M. We hypothesized that this population still includes patients who may benefit from osimertinib and are thus deprived of an effective therapy because of limitations of current biomarker assay technologies.
PET using the tracer 18F-FDG is an established staging modality in NSCLC and detects metabolically active tumor lesions. The term metabolic response describes a reduction in tumor metabolic activity by treatment as detected by sequential 18F-FDG PET scanning (9). In NSCLC, metabolic responses have been reported with cytotoxic chemotherapy (10), radiation therapy (11), and targeted therapies (12).
Against this background, we assessed whether a metabolic response by 18F-FDG PET after short-course osimertinib treatment identifies patients with EGFR p.T790M-negative resistance to EGFR TKI who still benefit from next-line osimertinib.
MATERIALS AND METHODS
The open-label, single-armed phase II study THEROS (NCT03810066) explored the feasibility of combining a short course of osimertinib treatment with sequential 18F-FDG PET scanning. The study enrolled patients with metastatic EGFR-mutant NSCLC experiencing radiologically confirmed acquired resistance to first- or second-generation EGFR TKI. All patients tested negatively for EGFR p.T790M by centrally performed analysis of circulating free DNA. Absence of the EGFR p.T790M mutation was also confirmed in a tissue rebiopsy whenever medically safe and feasible. At least 1 tumor lesion with visually clearly increased 18F-FDG uptake above the background level by baseline PET/CT scanning was required. The target lesion was defined as the lesion with the highest 18F-FDG uptake, with no restrictions regarding tissue or organ type (i.e., local recurrence, lymph node and organ metastases). Following PERCIST 1.0, the metabolic volume of a target lesion for calculating the SUVpeak corrected for lean body mass was 1 mL. Patients received a single course (28 d) of osimertinib (80 mg daily). The early metabolic response was evaluated per PERCIST (9) by a second 18F-FDG PET/CT scan, which per protocol was scheduled between days 15 and 28. Patients with a metabolic response continued taking osimertinib until progression or withdrawal of consent. Patients without a metabolic response were offered standard-of-care therapy (Supplemental Fig. 1A). The primary study endpoint was the rate of metabolic responses to osimertinib, which was assessed using a modified Simon 2-stage design. Methods are further detailed in the supplemental materials (13–19).
RESULTS
From May 2019 to June 2021, 23 patients were screened, and 14 patients (50% female; median age, 69.6 y [range, 50.6–81.8 y]) were enrolled at 2 sites (Supplemental Fig. 1B; Table 1). Screening failures were due to detection of EGFR p.T790M in the central liquid biopsy (n = 4), other exclusion criteria (n = 2), patient refusal, and rapid clinical deterioration (1 patient each). Of note, a suitable 18F-FDG–avid lesion could be detected in all screening 18F-FDG PET/CT scans. We report clinical follow-up as of January 2023, with a median follow-up time of 27 mo (range of censored overall survival events, 16.6–34.2 mo). Twelve of 14 patients were evaluable for metabolic response. Two patients progressed clinically before the planned 18F-FDG PET/CT evaluation and were taken off the study. PET/CT was performed after a median of 21.5 d (interquartile range, 2.75 d; range, 13–31 d). Five patients of the intention-to-treat population (36%) achieved metabolic responses and continued study therapy (Figs. 1 and 2). At 12 mo, all 5 metabolic responders (100%) were still on treatment (Fig. 3). Median progression-free survival in these patients continuing study therapy was 18.7 mo (4 progression-free survival events; 95% CI, 14.6 mo to not estimable; Kaplan–Meier plot in Supplemental Fig. 2A). The protocol allowed continuation of osimertinib beyond progression in patients who maintained clinical benefit. All 4 metabolically responding patients who eventually experienced a progression-free survival event per RECIST continued osimertinib (Fig. 3). With 3 patients still on treatment, the median time to treatment failure in metabolic responders has not been reached.
Patient Characteristics (n = 14)
Response according to PERCIST and molecular profile. (Top) Relative change in SUVpeak corrected for lean body mass (SULpeak) on days 15–28 of osimertinib treatment (n = 12). Two patients (patients 1 and 2) were not evaluable because of clinical progression before protocol-defined metabolic response assessment. Patient responses were categorized according to PERCIST as metabolic response (green), metabolically stable disease (yellow), and metabolically progressive disease (orange). (Bottom) Somatic gene mutations detected in tumor and liquid biopsies from each patient, including 2 patients (patients 1 and 2) with rapid disease progression not preceding second 18F-FDG PET (left). Color-coded boxes indicate genomic alterations detected in tissue-derived DNA and circulating free DNA of respective patient (blue = EGFR mutations; magenta = TP53, KEAP1, STK11 mutations; green = MET, EGFR, ERBB2 copy number gain; brown = KRAS p.G12C mutation). Lighter colors indicate alterations detected at low variant allele frequencies.
Representative 18F-FDG PET/CT images from 2 patients illustrating partial response (top) and progressive disease (bottom) per PERCIST. At baseline, patient 10 had SUVpeak corrected for lean body mass of 18.8, which was reduced to 7.2 (−33%) by 1 cycle of osimertinib, whereas patient 4 increased from 9.2 at baseline to 15.6 at second imaging time point (+82%). ID = identification number.
Individual treatment trajectories of study patients. Duration of each treatment line before study enrollment (left of vertical black line indicating day 0 of study entry), study treatment, and postprogression treatment (right of vertical black line) is represented on x-axis. Individual treatment lines are color-coded (green = first- and second-generation EGFR-TKI; blue = osimertinib [study treatment and treatment beyond progression]; yellow = chemotherapy; orange = chemoimmunotherapy; purple = immunotherapy). Dashed red vertical lines indicate timing of protocol-defined metabolic response assessment per 18F-FDG PET/CT. Within each patient trajectory, arrows indicate ongoing osimertinib treatment at data cutoff (study treatment or treatment beyond progression). Red vertical bars indicate individual time points of progressive disease per RECIST. Vertical black bars indicate patient death.
Postprogression biopsies in 2 patients revealed potential mechanisms of osimertinib resistance, thus confirming the selective pressure of PET response–informed treatment: the EGFR p.C797S resistance mutation (20) was identified in an adrenal metastasis from 1 patient progressing on osimertinib. Subsequently, this patient responded to gefitinib reexposure. Phenotypic transformation from adenocarcinoma to large cell neuroendocrine lung cancer was confirmed in another patient progressing on osimertinib, who was subsequently managed with chemoimmunotherapy.
Early assessment of metabolic response was highly predictive of survival outcomes (Fig. 4; Supplemental Fig. 2): metabolic responders clearly had superior overall survival (only 1 death at 41.5 mo, P = 0.011 compared with patients with stable or progressive disease per PERCIST). Metabolically stable disease per PERCIST selected a subgroup with an intermediate prognosis. Three patients in this group—all experiencing a minor reduction in metabolic activity on 18F-FDG PET/CT—continued osimertinib outside the study (median progression-free survival, 7.2 mo). Patients with metabolically progressive disease had poor outcomes (Fig. 4).
Early metabolic response by 18F-FDG PET/CT indicates clinical benefit from osimertinib in patients with unknown resistance mechanisms to EGFR TKI. Shown is overall survival from initiation of study treatment according to PERCIST metabolic response category. PMD = progressive metabolic disease; PMR = partial metabolic response; SMD = stable metabolic disease.
The presence of the EGFR p.T790M resistance mutation in circulating free DNA was excluded in all patients by centrally performed next-generation sequencing (21). In addition, biobanked blood samples from study patients were retrospectively analyzed using highly sensitive Avenio circulating free DNA technology (Roche). This revealed a complex genomic environment including potential resistance mechanisms such as copy number gains of MET, EGFR, and ERBB2, and KRAS and CDKN2A mutations. No metabolically responding patient exhibited mutations in TP53 and STK11 or in MET and ERBB2 copy number gain (Fig. 1; Supplemental Fig. 3). In a single metabolically responding patient, the EGFR p.T790M mutation was retrospectively detected at a very low variant allele frequency (variant allele frequency, 0.78%) as compared with the original activating EGFR exon 19 indel (variant allele frequency, 1.6%). Of note, oncogenic KRAS p.G12C mutations were detected at low variant allele frequencies (0.23%–1.5%) in 7 patients. Only one of these patients (variant allele frequency, 0.23%) achieved a PERCIST response to osimertinib. Interestingly, patients not achieving a metabolic response to osimertinib had comutations of TP53, STK11, and KEAP1, and copy number gains of MET, ERBB2, and EGFR. Also, KRAS mutations were detected at low variant allele frequencies.
DISCUSSION
The study THEROS provides proof of concept for the utility of early metabolic response evaluation by 18F-FDG PET/CT as a predictor of clinical benefit from sequenced EGFR TKI therapy in patients with EGFR-mutated NSCLC. Currently, targeted therapy of NSCLC is offered on the basis of predictive genomic biomarkers. Although the predictive specificity of this approach is strong for first-line therapy, a targeted approach to next-line therapy is less developed. Using the well-defined clinical setting of acquired resistance to first- or second-generation EGFR TKI, we identified a considerable risk of withholding effective targeted therapy to biomarker-negative patients. Early metabolic response by 18F-FDG PET/CT was highly predictive of a patient population benefitting from sequenced osimertinib despite not demonstrating the appropriate predictive genomic biomarker, EGFR p.T790M.
To our knowledge, this was the first prospective study to explore a theranostic approach toward personalizing next-line therapy in patients with EGFR-mutant NSCLC acquiring resistance to EGFR TKI treatment of unknown mechanism. We showed that patients achieving an early metabolic response under theranostic short-term osimertinib treatment may derive a dramatic long-term clinical benefit. A key limitation of this proof-of-concept study is its modest cohort size. The introduction of early PET response assessment in prospective clinical studies of next-line TKI therapies is required to establish this novel theranostic approach in clinical practice.
In post hoc analyses, we have characterized the genomic landscape of our study patients with EGFR TKI resistance of unknown mechanism. Interestingly, patients not achieving a metabolic response to osimertinib had comutations of TP53, STK11, and KEAP1 and copy number gains of MET, ERBB2, and EGFR. Also, KRAS mutations were detected at low variant allele frequencies. Because the functional significance of these findings is unknown, they are purely hypothesis-generating.
CONCLUSION
Early metabolic response per 18F-FDG PET after short-course osimertinib is a promising dynamic biomarker for personalization of next-line therapy in EGFR-mutated NSCLC with acquired resistance to EGFR TKI. Next to EGFR-mutated NSCLC, this predictive paradigm could be translated to further settings of NSCLC with dominant driver oncogenes, in which multiple, non–cross-resistant targeted agents are available.
DISCLOSURE
This study received grant support (ESR-16-12451) and study medication from AstraZeneca. The West German Cancer Center and the Center for Integrated Oncology are supported by Oncology Center of Excellence grants from German Cancer Aid (Deutsche Krebshilfe). The West German Cancer Center received federal and state funding as a partner site of the German Cancer Consortium. None of the funding sources were involved in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the article for publication. Martin Schuler reported fees for consulting from Amgen, AstraZeneca, Blueprint Medicines, Boehringer Ingelheim, Bristol Myers Squibb, GlaxoSmithKline, Janssen, Merck Serono, Novartis, Roche, Sanofi, and Takeda; honoraria for continuing medical education presentations from Amgen, Boehringer Ingelheim, Bristol Myers Squibb, Janssen, MSD, Novartis, Roche, and Sanofi; and research funding to University Hospital Essen from AstraZeneca and Bristol Myers Squibb. Sebastian Michels reported fees for consulting and honoraria for continuing medical education presentations from Janssen and Lilly and research funding to the University of Cologne from Bristol Myers Squibb, Janssen, Novartis, and Pfizer. Martin Metzenmacher reported fees for consulting from Amgen, Astra Zeneca, Bristol Myers Squibb, GlaxoSmithKline, Janssen, MSD, Novartis, Roche, and Takeda. Jürgen Wolf reported fees for consulting and honoraria for continuing medical education presentations from Amgen, AstraZeneca, Bayer, Blueprint, Bristol Myers Squibb, Boehringer Ingelheim, Chugai, Daiichi Sankyo, Janssen, Lilly, Loxo, Merck, Mirati, MSD, Novartis, Nuvalent, Pfizer, Roche, Seattle Genetics, Takeda, and Turning Point and research funding to the University of Cologne from Bristol Myers Squibb, Janssen, Novartis, and Pfizer. Alexander Drzezga reported fees for consulting and speaker honoraria from Siemens Healthineers, Sanofi, GE Healthcare, Biogen, Novo Nordisk, Invicro, Novartis/AAA, and Bayer Vital; research support from Siemens Healthineers, Life Molecular Imaging, GE Healthcare, AVID Radiopharmaceuticals, Sofie, Eisai, and Novartis/AAA; stock from Siemens Healthineers and Lantheus Holding; and a patent for 18F-JK-PSMA7 (patent no. EP3765097A1). Ken Herrmann reported fees for consulting and continuing medical education presentations from Bayer, Sofie Biosciences, SIRTEX, Adacap Curium, Endocyte, IPSEN, Siemens Healthineers, GE Healthcare, Amgen, Fusion, Immedica, Onkowissen.de, Novartis, Molecular Partners, ymabs, Aktis Oncology, Theragnostics, Pharma15, Debiopharm, AstraZeneca, and Janssen; nonfinancial support from ABX; and grants and personal fees from BTG. No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: Can early metabolic response evaluation by 18F-FDG PET/CT serve as a predictor of clinical benefit from sequenced TKI therapy in patients with EGFR-mutated NSCLC?
PERTINENT FINDINGS: 18F-FDG PET after short-course osimertinib treatment identified patients with unknown mechanisms of TKI resistance deriving dramatic clinical benefit from sequenced osimertinib.
IMPLICATIONS FOR PATIENT CARE: The study provides proof of concept that early metabolic response assessment by 18F-FDG PET can be used as a novel paradigm for personalization of multiple lines of targeted therapies in patients with driver oncogene-dependent NSCLC.
ACKNOWLEDGMENTS
We thank the patients and their families who participated in the THEROS trial and all staff involved in patient care, study conduct, and translational studies at University Hospital Essen, University Hospital Cologne, and the CRO, ClinAssess, Leverkusen. The West German Biobank Essen is acknowledged for supporting the exploratory biomarker analyses.
Footnotes
Published online Apr. 4, 2024.
- © 2024 by the Society of Nuclear Medicine and Molecular Imaging.
REFERENCES
- Received for publication November 6, 2023.
- Revision received February 26, 2024.