Visual Abstract
Abstract
The aim of this study was to validate a previously developed deep learning model in 5 independent clinical trials. The predictive performance of this model was compared with the international prognostic index (IPI) and 2 models incorporating radiomic PET/CT features (clinical PET and PET models). Methods: In total, 1,132 diffuse large B-cell lymphoma patients were included: 296 for training and 836 for external validation. The primary outcome was 2-y time to progression. The deep learning model was trained on maximum-intensity projections from PET/CT scans. The clinical PET model included metabolic tumor volume, maximum distance from the bulkiest lesion to another lesion, SUVpeak, age, and performance status. The PET model included metabolic tumor volume, maximum distance from the bulkiest lesion to another lesion, and SUVpeak. Model performance was assessed using the area under the curve (AUC) and Kaplan–Meier curves. Results: The IPI yielded an AUC of 0.60 on all external data. The deep learning model yielded a significantly higher AUC of 0.66 (P < 0.01). For each individual clinical trial, the model was consistently better than IPI. Radiomic model AUCs remained higher for all clinical trials. The deep learning and clinical PET models showed equivalent performance (AUC, 0.69; P > 0.05). The PET model yielded the highest AUC of all models (AUC, 0.71; P < 0.05). Conclusion: The deep learning model predicted outcome in all trials with a higher performance than IPI and better survival curve separation. This model can predict treatment outcome in diffuse large B-cell lymphoma without tumor delineation but at the cost of a lower prognostic performance than with radiomics.
- diffuse large B-cell lymphoma
- maximum-intensity projection
- convolutional neural networks
- time to progression
- prediction
A combination of 18F-FDG PET with CT imaging is the preferred imaging modality for staging in diffuse large B-cell lymphoma (DLBCL) patients (1). Because of the heterogeneity of the disease, the current first-line treatment strategy in DLBCL results in relapse in one third of patients within the first 2 y (2). The International Prognostic Index (IPI) is used in the clinic to estimate patient prognosis, but it has suboptimal performance (3). Different PET-derived metrics are reported to be strong prognostic factors for DLBCL, especially metabolic tumor volume (MTV) (4). These metrics can be incorporated into prediction models for patient prognosis, also known as radiomic models. However, tumor delineation is required for the extraction of these metrics, which can be labor-intensive and is prone to intra- and interreader variabilities. Thus, there is a need to develop user-independent, effective, and reliable methods that can aid the identification of high-risk DLBCL patients.
Artificial intelligence and deep learning are promising technologies in the field of medical imaging. Their clinical applications are vast and include diagnostics, postprocessing techniques, tumor detection and delineation, prognosis, and clinical decision-making (5). One of the main drawbacks of deep learning applied to medical imaging is the large computational requirements for the training of the models. PET scans are extremely large in terms of memory size, and thus the analysis of such inputs requires deep learning models with increasingly more complex layers. The use of maximum-intensity projections (MIPs) can mitigate this because PET scans are projected onto 2-dimensional images, largely reducing the computational burden (6). The advantage of deep learning over radiomic models is that the former can directly learn from the images, without the need for lesion delineation to enable prediction of disease progression. In contrast, radiomic models are based on tumor segmentations. There is increasing interest in the use of deep learning and convolutional neural networks (CNNs) in DLBCL, especially for automatic tumor segmentation (7). However, there are only a few studies that focus on the use of CNNs for prediction of tumor progression directly from segmentation-free PET images (8,9).
In a previous study, we developed a CNN for the prediction of 2-y time to progression (TTP) from coronal and sagittal MIP images of DLBCL baseline scans (i.e., MIP-CNN) (10). This model was trained on the clinical trial HOVON-84 (11) and was externally evaluated on an independent clinical trial, PETAL (12). Proper validation of such models is difficult, as the predictive performance varies across populations and target settings and can also change with, for example, time due to improvements in care (13). Therefore, external validation is an essential aspect of assessing model performance, especially with deep learning models, since the decision-making processes of these can be challenging to understand.
The aims of this study were to extend the validation of the MIP-CNN model to 5 other international clinical trials and to compare the predictive performance of the MIP-CNN model with the IPI score, which is the current clinical standard, and 2 other radiomic models that require tumor segmentation.
MATERIALS AND METHODS
Study Population
In total, 1,466 18F-FDG PET/CT baseline scans from newly diagnosed DLBCL patients were available, and after quality control, 836 scans were used in this study. Quality control exclusions are provided in the Results section. These patient scans were obtained from 6 independent clinical trials: PETAL (12), GSTT15 (14), IAEA (15), NCRI (16), SAKK (17), and HOVON-130 (18). Additionally, 296 patients from the HOVON-84 trial were used to train the models as reported previously (4,10). All patients were treated with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone with a varying number of cycles, mostly 6 or 8. Individual trials were approved by the institutional review board, and all patients provided written informed consent. The institutional review board of the VU University Medical Center (JR/20140414) approved the use of these data. Details on the quality control of the scans can be found in Supplemental Data 1 (supplemental materials are available at http://jnm.snmjournals.org).
Prediction Models
In this study, 3 different models were implemented for the prediction of risk of progression within 2 y from the time of the baseline scan (i.e., 2-y TTP): the MIP-CNN model developed by Ferrández et al. (10) (a deep learning model that uses coronal and sagittal MIP images as inputs), the clinical PET model developed by Eertink et al. (4), and a PET model that used PET features only. The MIP-CNN was previously trained on the HOVON-84 trial and initially tested on the PETAL trial (n = 340) (10). In this study, the MIP-CNN model was externally tested on an additional 5 clinical trials, accounting for 496 patients, whereas the newly proposed PET model was not tested using the PETAL data and was therefore externally tested on all 836 scans. Yet, to allow direct comparison, we report the results for both the MIP-CNN and the new PET model for all external trial datasets. For the definition of TTP, patients who died within 2 y from the time of the baseline scan without signs of progression were excluded from the analysis. An illustration of the model’s design can be found in Figure 1. The performance of the 3 models was compared with the IPI risk score. The IPI was established using low-, low-intermediate-, high-intermediate-, and high-risk groups (3).
Flowchart of steps involved in design of models. (A) MIP-CNN: MIP images are obtained from baseline PET scans and used as input of model. (B) Radiomic models: tumors are delineated for each patient, and features are extracted from these delineations. Those features are used as predictors in machine learning model. Clinical PET model includes clinical parameter as predictors (age and WHO status). All models are designed to predict probability of 2-y TTP.
MIP-CNN Model
MIP images were generated using an in-house–developed preprocessing tool in Interactive Data Language (IDL; NV5 Geospatial Solutions, Inc.). This tool produces coronal and sagittal MIPs with dimensions of 275 × 200 × 1 and a pixel size of 4 × 4 mm. Examples of these coronal and sagittal MIPs are illustrated in Figure 1A. Details on the design of the MIP-CNN are described in Supplemental Data 2 (10). The model is available for download from the supplemental files from an article by Ferrández et al. (10).
Radiomic Models
The tumors were delineated using an SUV threshold of 4.0, as recommended by Barrington et al. (19). and used previously by Eertink et al. (4). Additionally, any physiologic uptake included in tumor regions was manually removed, because of the proximity of the physiologic uptake to the tumor. All delineations were performed using the ACCURATE tool (20). More details on the delineation process and the extraction of the PET features were given in previous studies (21).
The clinical PET model was first developed using the HOVON-84 trial by Eertink et al. (4). The clinical PET model includes the following features: MTV, SUVpeak, maximum distance from the bulkiest lesion to another lesion, age, and World Health Organization performance status. These features were used as predictors in a logistic regression model to predict the probability of 2-y TTP for each patient.
The PET model followed the same design as the clinical PET model but included only PET-extracted features as predictors: MTV, SUVpeak, and maximum distance from the bulkiest lesion to another lesion.
Statistical Analysis
The receiver operator characteristic curve and the area under the curve (AUC) were used to evaluate the models’ performance. The radiomic models were internally validated with a stratified repeated 5-folds cross-validation (CV) (4). The internal validation of the MIP-CNN followed a 5-fold CV with a balancing data scheme, which was explained in detail previously (10). A 2-sided Delong test was used to assess the differences between models’ performance in terms of AUC (22). To externally test the radiomic models, the model coefficients were fixed and used to calculate the 2-y TTP probabilities. In the case of the MIP-CNN, the model weights were saved during training and later used to calculate the probabilities.
Kaplan–Meier analysis was used to obtain the survival curves for each model, and these were compared against IPI. We found that most events in this population occur during the first 2 y of treatment, and therefore, the data after this period are not included in the survival analysis. High-risk IPI is defined by the presence of 4–5 adverse factors. High-risk groups for the MIP-CNN, clinical PET, and PET prediction models were defined by the patients with the highest predicted probabilities. To facilitate comparison between the high-risk groups in the IPI and the prediction models, the high-risk patient cohort for the prediction models were matched in size to the high-risk IPI group.
To further evaluate and quantify the models’ predictive performance in terms of overall fit, calibration, and discrimination, we reported the calibration plot, the slope and intercept of the calibration, the Brier score (23), and the absolute average difference between observed and predicted probabilities for each model Supplemental Data 3 (13).
These analyses were conducted in R (version 4.3.2). A P value below 0.05 was considered statistically significant.
RELAINCE Artificial Intelligence Claim
In this paper, the externally validated artificial intelligence method was designed for predicting 2-y TTP, and it was trained and validated for baseline 18F-FDG PET/CT studies in DLBCL patients. The method was validated and tested for baseline PET studies in DLBCL patients only when PET data needed to comply with the image quality standards as previously outlined (10) and preferably applied to PET acquired following EARL standards 1. The performance of the method was evaluated using the AUC and Kaplan–Meier plots.
RESULTS
Study Population
The HOVON-84 dataset was used to train the models described in this study. In total, 296 HOVON-84 DLBCL patients were used to train the model. Details on exclusion and inclusion criteria were previously published (10).
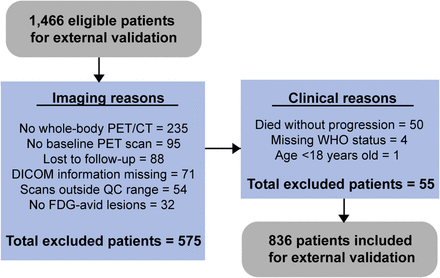
In total, 1,466 scans were available for external validation in the PETRA database. Exclusion criteria for this study included the absence of baseline 18F-FDG PET imaging (n = 95), no follow-up data within 2 y (n = 88), missing World Health Organization performance status (n = 4), and age below 18 y (n = 1). Quality control procedures resulted in the exclusion of patients with incomplete scans (n = 235), essential Digital Imaging and Communications in Medicine information missing (n = 71), no 18F-FDG–avid lesions (n = 32), and scans outside the quality control range (n = 54). Additionally, 50 patients died without progression within 2 y. This resulted in a total of 836 patients included in the study and used for external validation of the model as shown in Figure 2. A description of patient characteristics for all clinical trials is given in Supplemental Table 2.
Flowchart of selection of patients included in study for external validation. QC = quality control.
Prediction Models
The CV-AUC of the HOVON-84 trial yielded 0.67 for the IPI model. The MIP-CNN yielded a CV-AUC of 0.72, outperforming the IPI. Both the clinical PET and the PET models also outperformed IPI and the MIP-CNN, with CV-AUCs of 0.76 and 0.75, respectively. The SD, sensitivity, and specificity for each model are given in Table 1. The Youden index was used to establish the threshold for sensitivity and specificity.
SD, Sensitivity, and Specificity for Models’ AUCs
The external validation on all patients yielded an AUC of 0.60 for the IPI model, whereas the MIP-CNN achieved an AUC of 0.66, which was significantly higher than for the IPI model (P < 0.01). This was also the case for the radiomic models, both of which had significantly higher AUCs than did the IPI: 0.69 for the clinical PET model and 0.71 for the PET model (P < 0.001). This is illustrated in Figure 3. There were no statistical differences between the AUCs of the MIP-CNN and the clinical PET model; however, the PET model yielded a significantly higher AUC than did the MIP-CNN and clinical PET models (P < 0.05). An overview of all AUCs for each individual clinical trial can be found in Figure 4 and Supplemental Table 3. Receiver operating characteristic curves considering the 5 additional clinical trials only are shown in Supplemental Figure 3. As an additional exploratory analysis, we also tested the models’ performances to predict overall survival on all external data (Supplemental Fig. 4).
Receiver operating characteristic curves for 2-y TTP for all external data for 3 prediction models compared with IPI score.
AUCs of IPI, MIP-CNN, clinical PET, and PET prediction models for all 7 trials, including CV-AUC for training set (H84) and for all 6 external clinical trials together (ALL).
Patients classified as high-risk showed significantly reduced survival rates compared with those classified as low-risk across all prediction models (P < 0.0001). The Kaplan–Meier curves are illustrated in Figure 5. The survival rate for patients within the IPI high-risk group was 67.5%, with a 95% CI of 61.4–74.2. The survival rates for patients within the MIP-CNN, the PET, and the clinical PET high-risk groups were 61.1% (95% CI, 54.8–67.3), 57.1% (95% CI, 50.7–64.3), and 57.1% (95% CI, 50.7–64.3), respectively.
Kaplan–Meier survival curves of 2-y TTP stratified into low- and high-risk groups for IPI and MIP-CNN prediction models (A), IPI and clinical PET prediction models (B), and IPI and PET prediction models (C).
DISCUSSION
In this study, we evaluated the predictive performance of the MIP-CNN model in 5 independent clinical trials: GSTT15, IAEA, SAKK, NCRI, and HOVON-130. This model was previously developed and trained using the HOVON-84 trial and initially tested only on the PETAL trial (10). We have shown that the model remains predictive of outcome in all 6 independent clinical trials. Moreover, The MIP-CNN outperformed the IPI score when evaluated on all 836 external patients, and the performance remained consistently better than that of IPI for each individual clinical trial.
The use of U-nets and nnU-nets is increasingly growing, and their application on PET imaging for lymphoma is promising (24,25). Novel artificial intelligence–based methods in this field are mostly applied for tumor segmentation; however, there are only a few papers on the use of deep learning models for outcome prediction in DLBCL. To our knowledge, only 2 other studies have used CNNs with 18F-FDG PET images as the main input. Also using coronal MIPs, Rebaud et al. trained a multitask ranker neural network whose performance for progression-free survival prediction was equivalent to that of total MTV segmented by experienced nuclear medicine physicians (9). In contrast, Liu et al. (8), developed a 3-dimensional CNN for simultaneous automated lesion segmentation and prognosis prediction. These models show comparable performance to our MIP-CNN; however, unlike our study, these lack external validation and further assessment. Although there are similarities among our proposed CNN and the models used by Rebaud et al. and Liu et al., direct comparison of the 3 methods is hampered by the lack of information regarding model training and architecture in these 2 other studies. For this reason, we evaluated 2 radiomic model training procedures that could be replicated in the HOVON-84 trial. The clinical PET model, developed by Eertink et al., includes MTV, maximum distance from the bulkiest lesion to another lesion, SUVpeak, age, and World Health Organization performance status (4). The PET model is a simplified version of the latter, including only PET-extracted features (MTV, SUVpeak, and maximum distance from the bulkiest lesion to another lesion).
Another reason to include radiomic models in this study is the fact that they both require delineation of the tumors to extract prognostic information. The motivation behind this study was to build a prognostic model that would not require any delineation procedure, thus solving the issues that come with such tasks. Tumor delineation is time-consuming, ranging from 3 to 6 min per patient and up to 20 min for complicated cases (26). Moreover, there are no consensus guidelines on tumor delineation, and as a result, many different semiautomated methods are being used, providing substantially different PET uptake values and total MTVs (27).
When comparing the 3 models included in this study, we found that the radiomic models were associated with increased AUCs for 2-y TTP in all individual trials compared with the MIP-CNN. However, it is important to note that the MIP-CNN performance for the external validation on all 836 patients remained statistically equivalent to that of the clinical PET model. The performance differences between the MIP-CNN and the radiomic models may be explained by several factors. First, radiomic model predictors are extracted from manually curated tumor delineations. An SUV threshold of 4.0 is initially used to generate a tumor mask, which is then supervised by a nuclear medicine physician who edits the region to complete the final delineation. This may result in more accurate tumor delineation, with an experienced user removing adjacent physiologic uptake included in the mask and uptake likely due to causes other than lymphoma such as normal variants and infection or inflammation. Second, the use of MIP images instead of fully 3-dimensional images to build our deep learning model can have some limitations. Even though MIPs are more manageable and memory-efficient, it is possible that some relevant information is lost when using MIPs, possibly resulting in less precise predictions. Nevertheless, compared with the radiomic models, the MIP-CNN is a model free of tumor segmentation; therefore, it is easier to apply, allowing routine use in clinical practice.
The 3 models follow a similar trend in performance across the datasets, as seen in Figure 4. This trend might indicate that there are case-mix differences in the data between the clinical trials, affecting the model performance in a consistent way for all models. Those differences may arise from specific patient characteristics that are not considered when building the models. The HOVON-130 trial was the study that included only patients with MYC oncogene rearrangements, which is a well-known high-risk feature (18). This could explain the low AUCs achieved by the 3 prediction models as well as the IPI for this clinical trial. IPI also performed poorly for NCRI, SAKK, and IAEA, which all present a relatively higher proportion of low-risk patients than the other clinical trials. Moreover, SAKK and IAEA consist of a relatively younger population, especially compared with HOVON-84. For these 2 clinical trials, the clinical PET model, which includes age as a predictor, yielded the highest AUC; however, for NCRI, GSTT15, and PETAL, the PET model, without clinical predictors, performed best. These results suggest that inclusion of age and World Health Organization status may have an impact on the model’s prognostic power, also considering that the PET model achieved a significantly higher AUC when tested on all external data.
There were some limitations in this study. We assessed only 2-y TTP as the outcome parameter. TTP was chosen because progression-free and overall survival are influenced by age and because age-related comorbidities and life expectancy can affect outcome for older patients independently of their lymphoma. Moreover, this provided continuity with previous studies that have also reported TTP as the primary endpoint (4,10). Most patients included in this study received rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone treatment; however, variations in treatment regimens across studies were observed, including differences in the number of cycles and the degree of treatment intensification. Another limitation of the MIP-CNN is the lack of activation maps to explain the predictions because of the design of the CNN with 2 branches, as discussed before (10). Yet, by digitally ablating the lesions from the MIPs, it is possible to demonstrate the impact of the lesion on the predictions. This requires, however, tumor segmentation, and to this end the use of artificial intelligence–based tumor segmentations is of interest and currently being explored (24,28).
In summary, this is the first study—to our knowledge—to show the potential of CNNs for outcome prediction and its applicability on such an extensive cohort of 18F-FDG PET baseline DLBCL scans.
CONCLUSION
The MIP-CNN was predictive of outcome in 5 individual external DLBCL trials, with a higher performance than IPI. The PET model had comparable performance to the clinical PET model, which both rely on tumor delineations. Our MIP-CNN can predict treatment outcome in DLBCL without tumor delineation but at the cost of a slightly decreased prognostic performance compared with other delineation-dependent models.
DISCLOSURE
This work was financially supported by the Hanarth Fonds Fund and the Dutch Cancer Society (VU-2018-11648). The sponsor had no role in gathering, analyzing, or interpreting the data. Sally Barrington received departmental funding from Amgen, AstraZeneca, BMS, Novartis, Pfizer, and Takeda. Martine Chamuleau received financial support for the clinical trials from Celgene, BMS, and Gilead. Josée Zijlstra received financial support for clinical trials from Roche, Gilead, and Takeda. Pieternella Lugtenburg received financial support for clinical trials from Takeda and Roche. No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: Can we use a deep learning model to predict outcome in DBLCL on multiple independent datasets?
PERTINENT FINDINGS: The deep learning model previously developed in the HOVON-84 dataset remained predictive of outcome in 5 independent external datasets.
IMPLICATIONS FOR PATIENT CARE: Implementation of deep learning could automate treatment outcome prediction, replacing tumor segmentation and user dependencies.
Footnotes
Published online Oct. 3, 2024.
- © 2024 by the Society of Nuclear Medicine and Molecular Imaging.
REFERENCES
- Received for publication June 6, 2024.
- Accepted for publication September 9, 2024.