Visual Abstract
Abstract
[177Lu]Lu-PSMA-617 was approved by the U.S. Food and Drug Administration for patients with prostate-specific membrane antigen (PSMA)–positive metastatic castration-resistant prostate cancer (mCRPC). Since the time of regulatory approval, however, real-world data have been lacking. This study investigated the efficacy, safety, and outcome predictors of [177Lu]Lu-PSMA-617 at a major U.S. academic center. Methods: Patients with mCRPC who received [177Lu]Lu-PSMA-617 at the Johns Hopkins Hospital outside clinical trials were screened for inclusion. Patients who underwent [177Lu]Lu-PSMA-617 and had available outcome data were included in this study. Outcome data included prostate-specific antigen (PSA) response (≥50% decline), PSA progression-free survival (PFS), and overall survival (OS). Toxicity data were evaluated according to the Common Terminology Criteria for Adverse Events version 5.03. The study tested the association of baseline circulating tumor DNA mutational status in homologous recombination repair, PI3K alteration pathway, and aggressive-variant prostate cancer–associated genes with treatment outcome. Baseline PSMA PET/CT images were analyzed using SelectPSMA, an artificial intelligence algorithm, to predict treatment outcome. Associations with the observed treatment outcome were evaluated. Results: All 76 patients with PSMA-positive mCRPC who received [177Lu]Lu-PSMA-617 met the inclusion criteria. A PSA response was achieved in 30 of 74 (41%) patients. The median PSA PFS was 4.1 mo (95% CI, 2.0–6.2 mo), and the median OS was 13.7 mo (95% CI, 11.3–16.1 mo). Anemia of grade 3 or greater, thrombocytopenia, and neutropenia were observed in 9 (12%), 3 (4%), and 1 (1%), respectively, of 76 patients. Transient xerostomia was observed in 23 (28%) patients. The presence of aggressive-variant prostate cancer–associated genes was associated with a shorter PSA PFS (median, 1.3 vs. 6.3 mo; P = 0.040). No other associations were observed between circulating tumor DNA mutational status and treatment outcomes. Eighteen of 71 (25%) patients classified by SelectPSMA as nonresponders had significantly lower rates of PSA response than patients classified as likely responders (6% vs. 51%; P < 0.001), a shorter PSA PFS (median, 1.3 vs. 6.3 mo; P < 0.001), and a shorter OS (median, 6.3 vs. 14.5 mo; P = 0.046). Conclusion: [177Lu]Lu-PSMA-617 offered in a real-world setting after regulatory approval in the United States demonstrated antitumor activity and a favorable toxicity profile. Artificial-intelligence–based analysis of baseline PSMA PET/CT images may improve patient selection. Validation of these findings on larger cohorts is warranted.
- metastatic castration-resistant prostate cancer
- PSMA PET
- ctDNA
- SelectPSMA
- LuPSMA
- radiopharmaceutical therapy
The U.S. Food and Drug Administration approval of [177Lu]Lu-PSMA-617 (Pluvicto; Novartis) for patients with prostate-specific membrane antigen (PSMA)–positive metastatic castration-resistant prostate cancer (mCRPC) was based on the results from the VISION trial (1). Data on the real-world efficacy and toxicity profile of [177Lu]Lu-PSMA-617 in the period since its regulatory approval are scarce. The National Comprehensive Cancer Network guideline recommends that candidates for [177Lu]Lu-PSMA-617 receive [68Ga]Ga-PSMA-11, [18F]DCFPyL, or [18F]rh-PSMA-7.3 PET/CT (henceforth collectively referred to as PSMA PET/CT) to determine treatment eligibility. Despite the use of pretreatment PSMA PET/CT as a gatekeeper, only 46% of patients achieved a prostate-specific antigen (PSA) response in the VISION trial (1).
Over the past few years, efforts in the prostate cancer theranostics community have focused on developing advanced strategies to identify nonresponders using baseline tumor and patient characteristics (2,3) and to improve patient selection for PSMA-targeted therapies. Tumor uptake on PSMA PET imaging was proposed as a predictive biomarker for PSA response and as a prognostic biomarker for progression-free survival (PFS) and overall survival (OS) (2–4), but validation data are needed. The use of circulating tumor DNA (ctDNA) is becoming increasingly important as a noninvasive blood-based biomarker in metastatic prostate cancer (5). Preliminary reports have investigated the prognostic value of ctDNA for outcome after [177Lu]Lu-PSMA therapy, but the findings are heterogeneous and inconclusive (6–8).
The primary objective of this study was to evaluate the efficacy and safety profile of [177Lu]Lu-PSMA-617 in a real-world setting at a major U.S. academic center. The study’s secondary objective was to investigate factors associated with outcome after [177Lu]Lu-PSMA-617.
MATERIALS AND METHODS
This was a single-center, retrospective study. The institutional database was screened to identify patients with prostate cancer who received [177Lu]Lu-PSMA-617 at the outpatient center of the Johns Hopkins Hospital outside clinical trials. To receive [177Lu]Lu-PSMA-617, patients had to have mCRPC, have progressed on (or be unfit to receive) taxane-based chemotherapy and androgen receptor-signaling inhibitor (ARSI) agents, and have had PSMA-positive disease on baseline PSMA PET/CT imaging. The decision to treat patients was made by the local interdisciplinary care team, which involved urologists, medical oncologists, and nuclear medicine physicians. Patients eligible for this analysis had to receive at least 1 cycle of [177Lu]Lu-PSMA-617 if treatment was completed or at least 4 cycles of [177Lu]Lu-PSMA-617 if treatment was ongoing at the last follow-up and have outcome data available. Treatment was considered completed if patients received the maximum number of cycles or if the treatment was discontinued because of tumor progression or toxicity.
The Johns Hopkins Institutional Review Board approved this retrospective study, and the requirement to obtain informed consent was waived.
Procedures
All patients received baseline PSMA PET/CT with [68Ga]Ga-PSMA-11 or [18F]DCFPyL before [177Lu]Lu-PSMA-617 initiation. PSMA PET/CT images were evaluated by nuclear medicine specialists according to the VISION PET criteria to determine eligibility status for treatment (1).
Treatment with 7.4 GBq of [177Lu]Lu-PSMA-617 was administered every 6 wk for a maximum of 6 cycles, in the absence of progression or severe toxicity, according to the treating physician. Clinical laboratory assessments including serum PSA, complete metabolic panel, and complete blood count were performed every 6 wk. Before treatment with [177Lu]Lu-PSMA-617, a subset of patients underwent somatic or germline genomic panel testing as a standard-of-care procedure, using commercially available DNA-sequencing platforms (FoundationOne, Personal Genome Diagnostics, Color Health, or Invitae).
Outcome
Efficacy was determined by the PSA response rate (PSA50-RR), PSA PFS, and OS. PSA50-RR was defined as the percentage of patients who achieved at least a 50% PSA decline relative to baseline at any time during treatment. PSA PFS was defined as the time from treatment initiation to PSA progression or death, whichever occurred first, with PSA progression being defined according to Prostate Cancer Working Group Criteria 3 as at least a 25% increase and at least 2 ng/mL above the nadir (9). OS was defined as the time from treatment initiation to death from any cause or last follow-up alive. The cutoff date for the last follow-up was February 2, 2024.
The hematologic and nonhematologic safety profile was evaluated according to the Common Terminology Criteria for Adverse Events version 5.03. All adverse events that occurred after treatment initiation and within 12 wk of the last cycle of [177Lu]Lu-PSMA-617 were reported. Nephrotoxicity after [177Lu]Lu-PSMA-617 was evaluated at 3, 6, and 12 mo by estimated glomerular filtration rate (eGFR), using the Chronic Kidney Disease Epidemiology Collaboration creatinine formula, which is based on age and sex and is independent of race (10). Renal failure stages were subsequently defined according to Chronic Kidney Disease Epidemiology Collaboration criteria at baseline and at the 12-mo follow-up (11).
ctDNA Analysis
The study investigated the association of the presence of somatic or germline mutations before [177Lu]Lu-PSMA-617 with outcome data. Mutations in homologous recombination repair (HRR) genes (BRCA1, BRCA2, ATM, CHECK2, RAD51D, PALB2, NBN, BRIP1), PI3K pathway genes (PTEN, PIK3CA, PIK3CB, PIK3R1, AKT1), aggressive variant prostate cancer (AVPC)–associated genes (TP53, RB1, PTEN: a set of partially redundant tumor suppressor genes whose disruption is associated with an aggressive clinical course and heightened sensitivity to chemotherapy), the AR gene, and TMPRSS2-ERG gene fusion were tested. One alteration in HRR and PI3K pathway genes was sufficient to consider mutational status positive, whereas alterations in at least 2 AVPC genes were required for positive mutational status (12). We used the variant allelic fraction cutoff used by the sequencing vendor (0.4%–0.82% frequency).
PSMA PET/CT Image Analysis
Digital Imaging and Communications in Medicine data of baseline PSMA PET/CT images were available for 71 of 76 (93%) patients. Baseline PSMA PET/CT images were analyzed by a nuclear medicine physician using the NucsAI platform. The software was provided to investigators as a research tool to analyze PSMA PET/CT images using 2 artificial intelligence (AI) products (i.e., DeepPSMA and SelectPSMA). DeepPSMA and SelectPSMA had been previously developed by the company; the current dataset was not used for training or for validation of these technologies. The Digital Imaging and Communications in Medicine files of the PSMA PET/CT images were uploaded to the software platform and automatically analyzed by the AI.
DeepPSMA is a software product that automatically detects and segments tumors on PSMA PET/CT images. Once image analysis was completed, quantitative tumor burden parameters were automatically generated, including the whole-body SUVmean, which represents the average PSMA uptake of all tumor lesions. Patients were classified on the basis of a tumor SUVmean cutoff of 10 (SUV10), as described previously (2). The whole-body tumor SUVmean was also implemented in 177Lu-PSMA (LuPSMA) outcome classification models (3).
SelectPSMA is a software product that analyses PSMA PET/CT images to predict treatment responses to PSMA-targeted therapeutic radiopharmaceuticals. The technology performs advanced analyses of tumor lesions detected and annotated by DeepPSMA. SelectPSMA was used on the current dataset of baseline PSMA PET/CT images to classify patients as responders (PSMA-Rs) versus nonresponders (PSMA-NRs) to [177Lu]Lu-PSMA-617.
LuPSMA Nomograms
Development of original outcome-classification models (nomograms) was based on a multicenter study that retrospectively included 270 patients with mCRPC treated with [177Lu]Lu-PSMA-617 or [177Lu]Lu-PSMA-I&T at 6 medical centers (3). The endpoints of the nomograms were OS, PSA PFS, and PSA response; 1 nomogram was developed for each endpoint. The nomograms included the following disease history information for each patient: time since diagnosis of prostate cancer, previous chemotherapy status, baseline laboratory testing (hemoglobin level), and screening PSMA PET/CT parameters (whole-body tumor SUVmean, number of PSMA-positive metastases [<20 or >20], presence of pelvic lymph node metastases, presence of bone metastases, and presence of liver metastases) (Supplemental Table 1; supplemental materials are available at http://jnm.snmjournals.org). The original LuPSMA nomograms (3) were applied to the present cohort to obtain the individual risk scores. The previously established cutoffs for nomogram-obtained individual risk scores (41 points for PSA50-RR response, 178 for PSA PFS, and 197 for OS) were used to classify participants as having either a favorable outcome (higher probability for longer OS and PSA PFS and higher likelihood of PSA response) versus an unfavorable outcome (higher probability of shorter OS and PSA PFS and lower likelihood of PSA response).
Statistical Analysis
Results are presented as the median and interquartile range (IQR) for continuous variables and as the number and percentage for categoric variables. Kaplan–Meier analysis was used to calculate the PSA PFS and OS. A 2-sided Fisher exact test was used to evaluate the association of baseline ctDNA mutational status and PSMA PET-based predictions (SUV10, LuPSMA nomograms, and SelectPSMA) with PSA50-RR. The sensitivity, specificity, positive predictive value, and negative predictive value for SUV10, nomograms, and SelectPSMA to predict PSA response were calculated. Hazard ratios and the corresponding 95% CIs were used to evaluate the association of baseline ctDNA mutational status and PSMA PET-based predictions with PSA PFS and OS. Kaplan–Meier analysis and log-rank testing were performed to calculate and plot the association of PSMA PET-based parameters with PSA PFS and OS. A P value of less than 0.05 was considered statistically significant. Statistical analyses were performed using SPSS version 22 (IBM Corp.) and STATA version 18 (StataCorp LLC).
RESULTS
Patients
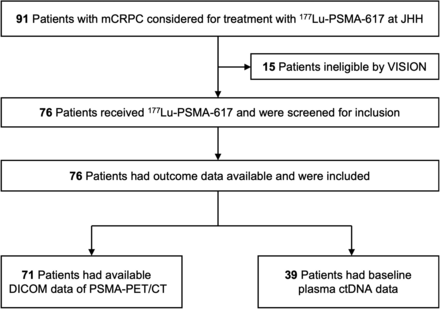
Of 91 consecutive patients screened for [177Lu]Lu-PSMA-617 treatment between December 1, 2021, and January 15, 2024, 76 (84%) met the VISION criteria and received the treatment. All 76 patients met the eligibility criteria for this retrospective analysis and were included (Fig. 1). Of the eligible patients, 70 (91%) completed the treatment, whereas 6 (8%) had ongoing treatment, receiving at least 4 cycles at the last follow-up. In total, 294 cycles were administered, with a median of 4 (IQR, 2–6) per patient. The median number of previous systemic treatments for mCRPC was 3 (IQR, 3–4). Seventy-three (96%) patients previously received taxane-based chemotherapy, and 76 (100%) received ARSIs. Fifty (66%) patients received 1 taxane regimen, and 23 (34%) received 2 taxane regimens. Thirty-eight (50%) patients received 1 ARSI regimen, and 38 (50%) received 2 ARSI regimens. Baseline patient characteristics are shown in Table 1. Forty-seven (62%) and 29 (38%) patients underwent baseline [18F]DCFPyL and [68Ga]Ga-PSMA-11 PET/CT, respectively. The median time from PSMA PET/CT to treatment initiation was 6.0 wk (IQR, 4.1–8.6 wk).
Consolidated Standards of Reporting Trials (CONSORT) flow diagram. DICOM = Digital Imaging and Communications in Medicine; JHH = Johns Hopkins Hospital.
Baseline Patient Characteristics (n = 76)
Efficacy
Two patients with undetectable PSA levels at baseline were not included in the PSA response analysis; 42 of 74 (57%) patients achieved any PSA reduction, and 30 (41%) achieved at least a 50% PSA reduction. Sixty-one (80%) patients had PSA progression at the last follow-up, and the median PSA PFS was 4.1 mo (95% CI, 2.0–6.2 mo). Thirty-seven (49%) patients had died by the last follow-up, and the median follow-up in survivors was 8.7 mo (IQR, 5.6–14.7 mo). The median OS was 13.7 mo (95% CI, 11.3–16.1 mo).
Safety Profile
Hematologic
Any grade of anemia, thrombocytopenia, leukopenia, neutropenia, or lymphopenia was observed in 19 (25%), 25 (33%), 30 (40%), 24 (32%), and 40 (53%) patients, respectively. Grade 3 or higher anemia, thrombocytopenia, neutropenia, leukopenia, or lymphopenia was observed in 9 (12%), 3 (4%), 1 (1%), 1 (1%), and 19 (25%) patients, respectively. Sixty-three (83%), 52 (68%), and 20 (26%) patients had available eGFR at 3, 6, and 12 mo, respectively. The median change in eGFR at 3, 6, and 12 mo relative to baseline was 0% (IQR, −4% to 7.9%), 0% (IQR, −6.9% to 4.3%), and −4.9% (IQR, −16.6% to 0%), respectively. At 12 mo, 5 of 20 (25%) patients had a clinically relevant moderate decrease in eGFR (≥15%) and 1 of 20 (5%) patients had a severe decrease (≥30%) in eGFR.
Nonhematologic
Grade 1 transient xerostomia was observed in 22 (28%) patients, fatigue in 10 (13%), nausea in 4 (5%), anorexia in 2 (3%), dysphagia in 1 (1%), and dry heaves in 1 (1%). Grade 2 xerostomia was observed in 1 (1%) patient. No treatment discontinuation or dose reduction of [177Lu]Lu-PSMA-617 due to adverse events was observed, and no treatment-related death occurred.
Outcome Prediction
Mutational Status
Thirty-nine of 76 (51%) patients underwent plasma ctDNA analysis before [177Lu]Lu-PSMA-617. Fourteen (36%), 15 (39%), 25 (64%), 9 (23%), and 16 (41%) patients had mutations in HRR, PI3K alteration pathway, AVPC, TMPRSS2-ERG, and AR-associated genes, respectively. The presence of AVPC was associated with a shorter PSA PFS (median, 1.3 vs. 6.3 mo; P = 0.040), and no other significant associations were observed between baseline mutational status by plasma ctDNA and treatment outcomes (PSA50-RR, PSA PFS, and OS) (Table 2).
Associations of ctDNA Mutational Status and PSMA PET Parameters with Outcome Measures
SUV10
Twenty-six of 71 (37%) patients had a whole-body tumor SUVmean of at least 10 on baseline PSMA PET/CT. These patients had a statistically higher PSA50-RR (56% vs. 30%, P = 0.04), longer PSA PFS (median, 7.0 vs. 3.3 mo; P = 0.046; Fig. 2A), and similar OS (median, 14.1 vs. 13.8 mo; P = 0.63; Fig. 2D) than patients with a tumor SUVmean of less than 10. The sensitivity, specificity, positive predictive value, and negative predictive value to detect patients with a PSA response were 52%, 74%, 56%, and 71%, respectively.
Survival curves by risk groups for PSA PFS stratified by SUV10 (A), PSA PFS nomogram (B), and SelectPSMA (C) and for OS stratified by SUV10 (D), OS nomogram (E), and SelectPSMA (F).
LuPSMA Nomograms
Fifteen (21%), 32 (45%), and 27 (38%) of 71 patients were classified as having an unfavorable outcome by PSA response, PSA PFS, and OS nomogram, respectively. Patients predicted to have an unfavorable outcome by nomograms had a significantly lower PSA50-RR (19% vs. 46%; P = 0.042), a shorter PSA PFS (median, 2.1 vs. 6.3 mo; P = 0.007; Fig. 2B), and a shorter OS (median, 6.3 vs. 16.1 mo; P < 0.001; Fig. 2E) than patients predicted to have a favorable outcome by nomograms. The sensitivity, specificity, positive predictive value, and negative predictive value to detect patients with a PSA response were 89%, 40%, 60%, and 81%, respectively. In a subanalysis, the presence of liver metastases was associated with a shorter PSA PFS (median, 1.3 vs. 4.8 mo; P = 0.020) and a shorter OS (median, 4.3 vs. 14.1 mo; P < 0.001).
SelectPSMA
Eighteen of 71 (25%) patients were classified as PSMA-NRs by the AI algorithm. PSMA-NRs achieved a significantly lower PSA50-RR (6% vs. 51%; P < 0.001) than PSMA-Rs. PSMA-NRs had a shorter PSA PFS (median, 1.3 vs. 6.3 mo; P < 0.001; Fig. 2C) and a shorter OS (median, 6.3 vs. 14.5 mo; P = 0.046; Fig. 2F) than PSMA-Rs. The sensitivity, specificity, positive predictive value, and negative predictive value to detect patients with a PSA response were 96%, 41%, 51%, and 94%, respectively. Of patients classified by LuPSMA nomograms as having an unfavorable outcome by PSA50-RR, PSA PFS, and OS, SelectPSMA classified 13 of 15 (87%), 13 of 32 (41%), and 8 of 27 (30%) as PSMA-NRs, respectively. PSMA-NRs had a lower PSA50-RR (0% vs. 40%; P = 0.045) and a shorter PSA PFS (1.2 vs. 3.0 mo; P = 0.002) than PSMA-Rs.
DISCUSSION
To the best of our knowledge, this study was the first since the regulatory approval of [177Lu]Lu-PSMA-617 to evaluate its efficacy and toxicity in a real-world setting in the United States. The analysis found a treatment outcome similar to real-world data reported in Germany before drug approval (13): a PSA50-RR of 40% versus 38%, a median PSA PFS of 4.1 versus 4.1 mo, and a median OS of 13.7 versus 12.9 mo. The safety profile was also similar in the present cohort versus the German study (13): grade 3 or higher anemia in 12% versus 9%, thrombocytopenia in 3% versus 4%, neutropenia in 1% versus 6%, and transient xerostomia in 23% versus 24%. No significant changes in renal function were observed in the first 6 mo after commencing [177Lu]Lu-PSMA-617 (median eGFR change, 0%). Nephrotoxicity rates at 12 mo were lower in the present cohort than in a previous report (11): at 12 mo, 25% versus 45% of patients had a moderate decrease in eGFR, and 5% versus 24% of patients had a severe decrease in eGFR. Of note, the current data are insufficient to establish a causal relationship between the administration of radiopharmaceutical therapies and renal impairment. Renal function in prostate cancer patients naturally declines as they approach death. Dedicated studies with a randomized design and an appropriate control group of patients receiving other therapeutic agents are required to provide definitive evidence on the long-term effects of [177Lu]Lu-PSMA-617 on kidney function. Such studies are particularly important because [177Lu]Lu-PSMA-617 is currently being investigated in patients in the earlier stages of prostate cancer, when life expectancy is longer (14,15).
The analysis also investigated predictive factors for outcome after [177Lu]Lu-PSMA-617. First, the mutational status by ctDNA at baseline (AVPC, HRR, PI3K pathway, AR, and TMPRSS2-ERG fusion genes) was tested for associations with outcome. Alterations in AVPC genes were significantly associated with PSA PFS but not with PSA50-RR or OS. Several small, retrospective studies previously evaluated associations between baseline ctDNA and outcome after [177Lu]Lu-PSMA-617, but the results were heterogeneous. Crumbaker et al. found associations of AVPC, PI3K pathway, TP53, and TMPRSS2-ERG genes with survival outcome (6). Vanwelkenhuyzen et al. identified associations of AR and PI3K pathway genes, but not TP53 and TMPRSS2-ERG, with survival outcome (7). In both studies, none of the molecular biomarkers correlated with PSA response. In contrast, another study found correlations of gene amplifications and CDK12 mutations with PSA response (8). Notably, the present study—as well as available data in the literature—is based on small patient cohorts, and the findings are inconclusive. Larger multicentric studies are required to definitively conclude whether the tumor genetic profile by ctDNA is associated with outcome after [177Lu]Lu-PSMA-617.
Second, the study explored previously proposed baseline PSMA PET-derived biomarkers for outcome after [177Lu]Lu-PSMA-617 (i.e., SUV10 (2) and LuPSMA nomograms) (3). This analysis confirmed the association of tumor SUV10 with PSA response (P = 0.04) and PSA PFS (P = 0.04) but not with OS (P = 0.63). The findings are in contrast to the original results, which found significant associations with OS in the TheraP trial data (4). The discrepant results may be explained by differences in the eligibility criteria used for [177Lu]Lu-PSMA-617 (i.e., patients underwent PSMA PET/CT at baseline in the current study, vs. dual-tracer PSMA and [18F]FDG PET/CT in the TheraP study) (4). SUV10 was proposed as a predictive biomarker for PSA response; however, patients with a tumor SUVmean of less than 10 who received [177Lu]Lu-PSMA-617 in the TheraP trial had a higher PSA50-RR than those who received cabazitaxel (52% vs. 36%) (2). The clinical value of SUV10 for patient selection is yet to be determined. Of note, SUV10 represents the average PSMA uptake of all tumor lesions (SUVmean), versus SUVmax, which provides the maximum PSMA uptake within tumor lesions.
Third, the study tested an AI-based algorithm that uses baseline PSMA PET/CT images to predict the response after [177Lu]Lu-PSMA-617. Patients classified as PSMA-NRs by SelectPSMA had a worse PSA50-RR (P < 0.001), PSA PFS (P < 0.001), and OS (P = 0.046) than patients classified as PSMA-Rs. PSMA-NRs had a median time to PSA progression of 1.3 mo (6 wk) after treatment initiation, suggesting that PSMA-NRs will likely progress by serum PSA after the first dose of [177Lu]Lu-PSMA-617. PSA progression after the first dose of [177Lu]Lu-PSMA radiopharmaceutical therapy was shown to be associated with a short radiographic PFS and OS (16), which underpins the prognostic value of SelectPSMA for long-term outcome. Further, a PSA flare phenomenon during [177Lu]Lu-PSMA radiopharmaceutical therapy is uncommon (16).
Patients classified as PSMA-NRs by both LuPSMA nomograms and SelectPSMA had worse short-term outcomes than patients classified as PSMA-NRs only by LuPSMA nomograms. These preliminary data suggest a potential added value of SelectPSMA over the nomograms in identifying PSMA-NRs to 177Lu-PSMA-617; however, prospective validation of these findings in larger datasets is required for definitive evidence.
Compared with SUV10 and LuPSMA nomograms, SelectPSMA achieved the highest negative predictive value (70% vs. 81% vs. 94%, respectively), suggesting it to be the most sensitive test for identifying patients who will not achieve a PSA reduction greater than 50%. The PSA50-RR for all patients treated with cabazitaxel is 36% (17), versus the lower PSA50-RR of 6% achieved by PSMA-NRs who received [177Lu]Lu-PSMA-617. A biomarker study is warranted to investigate whether patients classified as PSMA-NRs have a higher benefit from receiving cabazitaxel than from receiving [177Lu]Lu-PSMA-617. The main limitations of this study include its retrospective nature and single-center design. Validation of the findings in larger, multicentric patient cohorts is warranted.
CONCLUSION
[177Lu]Lu-PSMA-617 showed antitumor activity and a favorable toxicity profile in a real-world setting in patients with PSMA-positive mCRPC previously treated with taxane-based chemotherapy and ARSIs. Baseline ctDNA mutational status was not associated with treatment outcome. An AI algorithm, SelectPSMA, was the most accurate screening test to identify patients with the lowest likelihood of achieving at least a 50% PSA decline and patients with the highest risk of progression after [177Lu]Lu-PSMA-617, whereas LuPSMA nomograms were the most accurate at identifying patients with the highest risk of death. The clinical value of a tumor SUVmean of 10 is unclear. Validation of these findings in larger, multicentric prospective studies is warranted.
DISCLOSURE
Andrei Gafita was supported by the Prostate Cancer Foundation (21YOUN18). Channing Paller was supported by DOD W81XWH-22-2-0024 and P30CA006973; has a consulting role with Bayer, Dendreon, Omnitura, Exelixis, and AstraZeneca; and receives research funding from Lilly. Steven Rowe is a consultant for, and has received research funding from, Progenics Pharmaceuticals, Inc., a wholly owned subsidiary of Lantheus and the licensee of 18F-DCFPyL. Martin Pomper is a coinventor on a U.S. patent covering 18F-DCFPyL and as such is entitled to a portion of any licensing fees and royalties generated by this technology. This arrangement has been reviewed and approved by the Johns Hopkins University in accordance with its conflict-of-interest policies. No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: What is the efficacy, safety profile, and factors predictive of outcome for [177Lu]Lu-PSMA-617 in a real-world setting in the United States since regulatory approval of the drug?
PERTINENT FINDINGS: [177Lu]Lu-PSMA-617 showed antitumor activity and a favorable toxicity profile for patients with PSMA-positive mCRPC in a real-world setting. The baseline ctDNA mutational status was not associated with treatment outcome. Of the baseline PSMA PET/CT biomarkers, the SelectPSMA AI algorithm showed higher accuracy than a tumor SUVmean of 10 and LuPSMA nomograms in identifying PSMA-NRs to [177Lu]Lu-PSMA-617.
IMPLICATIONS FOR PATIENT CARE: Use of SelectPSMA may enhance patient selection for [177Lu]Lu-PSMA-617.
Footnotes
Published online Sep. 19, 2024.
- © 2024 by the Society of Nuclear Medicine and Molecular Imaging.
REFERENCES
- Received for publication March 4, 2024.
- Accepted for publication August 7, 2024.