Visual Abstract
Abstract
Evaluation of metabolic tumor volume (MTV) changes using amino acid PET has become an important tool for response assessment in brain tumor patients. MTV is usually determined by manual or semiautomatic delineation, which is laborious and may be prone to intra- and interobserver variability. The goal of our study was to develop a method for automated MTV segmentation and to evaluate its performance for response assessment in patients with gliomas. Methods: In total, 699 amino acid PET scans using the tracer O-(2-[18F]fluoroethyl)-l-tyrosine (18F-FET) from 555 brain tumor patients at initial diagnosis or during follow-up were retrospectively evaluated (mainly glioma patients, 76%). 18F-FET PET MTVs were segmented semiautomatically by experienced readers. An artificial neural network (no new U-Net) was configured on 476 scans from 399 patients, and the network performance was evaluated on a test dataset including 223 scans from 156 patients. Surface and volumetric Dice similarity coefficients (DSCs) were used to evaluate segmentation quality. Finally, the network was applied to a recently published 18F-FET PET study on response assessment in glioblastoma patients treated with adjuvant temozolomide chemotherapy for a fully automated response assessment in comparison to an experienced physician. Results: In the test dataset, 92% of lesions with increased uptake (n = 189) and 85% of lesions with iso- or hypometabolic uptake (n = 33) were correctly identified (F1 score, 92%). Single lesions with a contiguous uptake had the highest DSC, followed by lesions with heterogeneous, noncontiguous uptake and multifocal lesions (surface DSC: 0.96, 0.93, and 0.81 respectively; volume DSC: 0.83, 0.77, and 0.67, respectively). Change in MTV, as detected by the automated segmentation, was a significant determinant of disease-free and overall survival, in agreement with the physician’s assessment. Conclusion: Our deep learning–based 18F-FET PET segmentation allows reliable, robust, and fully automated evaluation of MTV in brain tumor patients and demonstrates clinical value for automated response assessment.
In recent years, several studies have demonstrated the clinical potential of volumetric response assessment in patients with brain tumors, particularly since the development of artificial neural networks has enabled this laborious task to be conducted in a fully automated way and with quality comparable to an experienced physician performing manual volumetry (1–3). For example, Kickingereder et al. (4) demonstrated the superior performance of an artificial neural network for the assessment of response to bevacizumab plus lomustine therapy for glioma patients based on structural MRI compared with the response assessment performed by a physician based on the Response Assessment in Neuro-Oncology criteria. The full integration of this method into the clinical workflow and the complete automatization allow for a more efficient, standardized, and reproducible volumetric evaluation of tumor burden, yielding great potential for response assessment in future clinical trials.
Although the clinical importance of structural MRI for response assessment is undisputed, there are known limitations for differentiation between treatment-related changes and tumor progression and for delineation of tumor extent, especially in cases of nonenhancing tumor portions (5–7). Because of its ability to overcome these shortcomings, amino acid PET has become an important diagnostic tool in patients with brain tumors. Specifically, amino acid PET is recommended by the Response Assessment in Neuro-Oncology group for response assessment in glioma patients at all disease stages (8,9). Among the amino acid PET tracers for patients with brain tumors, O-(2-[18F]fluoroethyl)-l-tyrosine (18F-FET) is the most widely used and evaluated PET tracer in Europe but is also gaining international importance, especially in the United States.
A prospective study conducted by Suchorska et al. on 79 patients with newly diagnosed glioblastoma showed that the metabolic tumor volume (MTV) assessed by 18F-FET PET before initiation of temozolomide chemoradiation was a strong prognostic factor for progression-free and overall survival, independent of the extent of resection (10). Recently, Ceccon et al. (11) found that in contrast to the MRI-based response assessment according to Response Assessment in Neuro-Oncology criteria and tumor-to-brain ratios (TBRs) for 18F-FET PET evaluation, MTV changes were predictive for the early identification of metabolic responders in patients undergoing adjuvant temozolomide chemotherapy. Furthermore, Wollring et al. (12) showed that MTV changes on 18F-FET PET are also an important factor for predicting response to lomustine-based chemotherapy in patients with recurrent gliomas. Despite these interesting findings, 3-dimensional assessment of MTV is not part of the routine clinical evaluation of amino acid PET, which is based mainly on TBR extracted from manually or semiautomatically generated 2-dimensional regions of interest (13). The fact that MTV is not routinely assessed in clinical practice suggests that the time and effort required for volumetric amino acid PET segmentation still exceed the clinical benefit. The number of studies investigating the clinical value of amino acid PET MTV needs to increase to demonstrate its clinical value and ultimately lead to the inclusion of volumetric amino acid PET assessment in consensus guidelines and recommendations.
To foster the clinical translation of volumetric amino acid PET evaluation, our study aimed to develop and evaluate an artificial neural network using the self-configuring no new U-Net (14) for the automated 3-dimensional segmentation of brain tumors using 18F-FET PET. Furthermore, the network was applied to a recently published 18F-FET PET study on response assessment in newly diagnosed glioblastoma patients treated with adjuvant temozolomide chemotherapy (11) for a fully automated response assessment in comparison to an experienced physician.
MATERIALS AND METHODS
Detailed methods can be found in the supplemental materials (available at http://jnm.snmjournals.org) (11,13–22).
Ethics
The study adhered to the standards established in the Declaration of Helsinki. The local ethics committees approved the retrospective analysis of imaging data (EK 055/19). All patients provided written informed consent before each 18F-FET PET investigation.
Patient Characteristics
Our database comprising 4,381 patients who underwent diagnostic 18F-FET PET scans at initial diagnosis, suspected tumor relapse, or treatment response assessment in our institution between November 2005 to April 2021 was retrospectively evaluated in this study. Of these 18F-FET PET scans, only those for which segmentations of MTV were available were included. Further, to evaluate the performance of the segmentation algorithm in patients lacking an increased 18F-FET uptake, 59 patients with iso- or hypometabolic 18F-FET PET scans were added. In total, 699 18F-FET PET scans from 555 patients were investigated in the study. Detailed patient characteristics are presented in Table 1.
Patient Characteristics
Data Sharing
The dataset, including 18F-FET PET image data and segmentations, is available on request. In addition, data analysis scripts in Python are available on request. The trained network (JuST_BrainPET) is available at https://github.com/MIC-DKFZ/nnUNet/tree/nnunetv1#useful-resources.
RESULTS
18F-FET Uptake Characteristics
Of the 476 18F-FET PET scans in the training dataset, 20 (4%) showed no pathologic uptake, 20 (4%) showed multifocal lesions, and 49 (10%) showed increased uptake due to nonmalignant lesions, for example, treatment-related changes. Of the 223 18F-FET PET scans in the test dataset, 39 (17%) showed no pathologic uptake, 19 (9%) showed multifocal lesions, and 26 (12%) showed nonmalignant lesions.
Network Performance for Lesion Detection
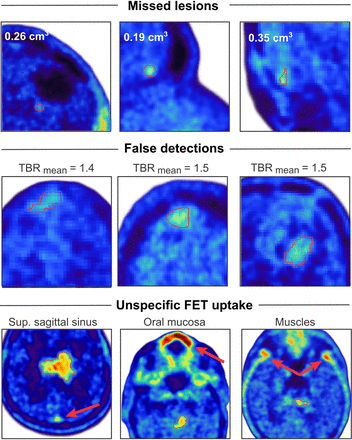
Of the 205 lesions with increased 18F-FET uptake, 189 were correctly identified by the network. Of 39 scans without increased uptake, only 6 were erroneously considered to show tumors by the network. Importantly, none of the anatomic regions that showed a physiologically increased uptake, such as in the superior sagittal sinus, were considered to be tumors by the network. This resulted in a mean F1 score of 92%, a sensitivity of 93%, and a positive predictive value of 95% for lesion detection. Patient examples showing lesions missed by the network, false detections, and examples of regions showing physiologically increased uptake are provided in Figure 1.
Network performance for lesion detection: ground truth segmentations of lesions that have not been detected by network, nonmalignant lesions with slightly increased but not pathologic uptake (mean TBR < 1.6) that have been erroneously detected as malignant lesions by network, and anatomic regions that show physiologically increased uptake that have always been correctly identified as such by network. TBRmean = mean TRB.
Network Performance for Lesion Segmentation
The median tumor volume was 11.1 cm3 (range, 0.03–109.4 cm3) for the training set and 10.6 cm3 (range, 0.1–98.8 cm3) for the test set (Table 1). In the training set, the mean volume Dice similarity coefficient (DSC) during 5-fold cross validation was 0.75 ± 0.03, and the mean surface DSC was 0.87 ± 0.03 without prior brain extraction. In the test set, the median volume DSC was 0.81 (interquartile range, 0.70–0.89), and the surface DSC was 0.96 (interquartile range, 0.89–0.99). With prior brain extraction, the mean volume DSC in the training set after 5-fold cross validation was 0.74 ± 0.03, and the mean surface DSC was 0.85 ± 0.02. In the test set, the median volume DSC was 0.80 (interquartile range, 0.68–0.88), and the median surface DSC was 0.93 (interquartile range, 0.87–0.98). Since brain extraction had no statistically significant effect on network performance (P > 0.05), the results in the following are based on the network trained without prior brain extraction. The network performance is summarized in Table 2. Some representative examples of tumor segmentations yielding low and high volume DSC and surface DSC are presented in Figure 2.
Performance of Network in Training and Test Datasets
Representative examples of lesion segmentations with high and low volume DSC. COMB = combination of ground truth and network segmentation; GT = ground truth segmentation; PRED = segmentation predicted by network; S-DSC = surface DSC; V-DSC = volume DSC.
The volume DSC and surface DSC were lowest for small lesions with a volume of between 0.1 and 3.3 cm3, which is equivalent to the first quartile of lesion volumes (median volume DSC, 0.65; interquartile range, 0.50–0.78; median surface DSC, 0.92; interquartile range, 0.78–0.99). For lesions of the second and third quartiles of lesion volumes (volume, 3.3–22.0 cm3), the median volume DSC was 0.80 (interquartile range, 0.71–0.88), and the median surface DSC was 0.93 (interquartile range, 0.88–0.99). The network showed the best performance for lesions from the fourth quartile of lesion volumes with a volume of between 22.0 and 98.0 cm3 (median volume DSC, 0.87 [interquartile range, 0.83–0.90]; median surface DSC, 0.97 [interquartile range, 0.94–0.99]).
Lesions with a larger MTV showed relatively low discrepancies between the predicted and the ground truth segmentations, compared with lesions with a smaller MTV (Fig. 3). This finding is also supported by a slight bias of the network in oversegmenting smaller MTVs, for example, the number of false-positive voxels was higher than that of false-negative voxels in smaller MTVs.
Absolute (A) and relative (B) differences between ground truth and predicted MTV of test dataset. Q1–Q4 = quartiles 1–4.
The number of false-positive voxels segmented by the network for the first, second/third, and fourth quartiles of lesion volumes was 65%, 21%, and 11%, respectively, and the number of false-negative voxels was 28%, 22%, and 15%, respectively. Single lesions were segmented with a better performance than nonmalignant and multifocal lesions (median volume DSC: 0.83, 0.77, and 0.67, respectively; median surface DSC: 0.96, 0.93, and 0.81, respectively) (Supplemental Fig. 3).
Automated Versus Manual Response Assessment
The 18F-FET PET parameter mean TBR extracted by the network was 2.1 ± 0.2 at baseline and 2.1 ± 0.2 at follow-up. The 18F-FET PET parameter mean TBR as evaluated by the physician was 2.0 ± 0.2 at baseline and 2.0 ± 0.2 at follow-up. The network and the physician agreed well in the assessment of MTV and in the clinical 18F-FET PET parameter mean TBR for both the baseline and the follow-up scans, with correlation coefficients ranging from 0.81 to 0.95 (Fig. 4).
Correlation between manual and automatic assessment of MTV (A) and mean TBR (B).
The 33 patients (median age, 50 y; range, 20–79 y; 17 women) had a median progression-free survival of 10 mo (range, 4–54 mo) and a median overall survival of 14 mo (range, 5–54 mo). The predicted baseline median MTV was 8.0 cm3 (range, 0.6–84.0 cm3), compared with a predicted follow-up median MTV of 12.6 cm3 (range, 0.6–121.4 cm3). The manually segmented median MTVs were 13.3 cm3 (range, 0.6–103.2 cm3) at baseline and 15.2 cm3 (range, 0.6–137.1 cm3) in the follow-up scans.
The network identified any decrease in MTV after temozolomide chemoradiation as an independent predictor for a significantly longer overall survival in glioma patients (P < 0.05). Relative changes in other parameters showed no significant predictive capability for a longer progression-free survival or overall survival. These findings were in line with the manual response assessment performed by an experienced physician. The corresponding Kaplan–Meier curves for progression-free survival and overall survival, along with representative 18F-FET PET images of patients with favorable and unfavorable prognoses, are shown in Figures 5 and 6.
Comparison of Kaplan–Meier curves for progression-free survival and overall survival assessed automatically by network and manually by experienced physician on basis of changes in mean TBR (A and C) and MTV (B and D).
Representative 18F-FET PET images at baseline and follow-up of glioma patients with favorable (top row) and unfavorable (bottom row) outcomes after 2 cycles of adjuvant temozolomide. OS = overall survival; PFS = progression-free survival; TBRmean = mean TRB.
DISCUSSION
The main finding of our study is that our deep learning–based neural network allows reliable and fully automated detection and 3-dimensional segmentation of brain tumors investigated by 18F-FET PET. Furthermore, the network demonstrated its clinical value for a fully automated 18F-FET PET assessment of response to temozolomide chemoradiation in glioma patients, whereby the network yielded results similar to the manual assessment performed by an experienced physician. This finding highlights the value of the network for improvement and automatization of clinical decision-making based on the volumetric evaluation of amino acid PET.
Currently, only a single study has investigated deep learning–based segmentation of brain tumors in adults using 18F-FET PET. Blanc-Durand et al. (23) demonstrated the potential of a 3-dimensional U-Net convolutional neural network for the automated detection of gliomas. Although the network achieved a comparable volume DSC of 0.79 in the validation set, the dataset comprised only a small number of patients (n = 37). Hence, the generalizability and clinical applicability of this approach remain questionable and require further verification.
The network developed in our study was able to correctly detect most tumors in the test dataset, resulting in high diagnostic performance (F1 score, 92%). Importantly, these results were obtained from a dataset that, in addition to patients with brain tumors and increased uptake, included patients with nonmalignant lesions that showed only slightly increased uptake, patients with no increased uptake, and even patients with photopenic defects (24).
Our network erroneously detected and segmented 6 of 39 nonmalignant lesions, for example, treatment-related changes, which showed a slightly increased uptake with a mean TBR of 1.5, which is just below the threshold of 1.6 that was used to generate the ground truth segmentations (Fig. 3B). Identifying these lesions unequivocally on the basis of 18F-FET PET imaging alone is a major challenge even for experienced nuclear medicine physicians, a fact that should be considered when evaluating the performance of our network. Furthermore, the lesions that were not correctly detected by the network were relatively small, with a mean MTV of 0.3 cm3. Hence, it seems that our network detected and segmented larger lesions more accurately (Fig. 3). This possibility is also supported by a slight bias of the network toward oversegmenting smaller MTVs; for example, the number of false-positive voxels was higher than the number of false-negative voxels in smaller MTVs, a fact that was already described by Blanc-Durand et al. (23).
These findings are in line with a recent study from Ladefoged et al. (25) in which an artificial neural network was developed and trained on 18F-FET PET and MRI scans from 233 adult brain tumor patients and applied to a dataset of 66 pediatric brain tumor patients for automated tumor segmentation. The authors also found the largest relative errors for tumor segmentations for small tumors with a volume of less than 10 cm3. Although the network demonstrated excellent performance in pediatric tumor patients, a few cases were reported in which the network erroneously delineated anatomic regions showing a high physiologic uptake. Such was not the case in our study, possibly because of the much larger number of patients used for training and the fact that our network was trained and evaluated on 18F-FET PET data from adults.
Of note, the fact that Ladefoged et al. (25) also included contrast-enhanced T1-weighted MR images as input images for the network might have had a positive effect on model performance. Since standardized anatomic MRI data were available for only a subset of patients in our study, we preferred to use a larger number of patients and omitted the addition of MRI. Nevertheless, the influence of the addition of MRI data should be investigated in future studies.
Another important finding of our study is the successful application of our fully automated 18F-FET PET tumor segmentation for the assessment of response in glioma patients after temozolomide chemoradiation. Similar to the manual response assessment performed by an experienced physician, our network also showed that a decrease in MTV was associated with a favorable outcome (Figs. 5 and 6). Beyond MTV, the evaluation of conventional 18F-FET PET parameters, especially TBRs, already plays an important role in the assessment of treatment response in clinical routine (8). In our study, we found a strong correlation in TBRs between the network and the manual assessment (Fig. 4).
The retrospective evaluation of our database revealed that MTV segmentation is still performed predominantly 2-dimensionally because of lack of 3-dimensional methods for the clinic. Availability of an automated method for 3-dimensional segmentation of MTV suitable for daily clinical use should therefore be in demand. Our network performs fully automated 3-dimensional segmentation of a single 18F-FET PET scan on a conventional graphics processing unit–equipped computer in less than 2 min without preprocessing, suggesting its suitability for successful implementation into clinical routine.
One limitation of our study is the uncertainty of the ground truth segmentation. Even though the segmentations were carefully performed according to the current guidelines for the evaluation of amino acid PET in brain tumor patients, interrater variability cannot be excluded. Nonetheless, this limitation is inherent in all work on segmentation and can hardly be overcome. Yet, ground truth uncertainties should be considered when the performance of a segmentation algorithm is being evaluated. A potential source of bias is patient selection, which was limited to patients for whom volumetric tumor segmentation was already available, rather than patients from a random sampling of a larger cohort, as might become possible if an automated 3-dimensional method of MTV segmentation were available. Another limitation—the low spatial resolution of PET—has a direct impact on the quality of the segmentations. To partly account for this limitation, development of our network was based on routinely acquired 18F-FET PET data from 2 PET scanners with different spatial resolutions. In the future, the addition of structural MRI might offer ways to minimize this effect. A further limitation might be that the network was trained on only 18F-FET PET data; its value for other commonly used amino acid PET tracers remains to be evaluated.
A general limitation is the comparatively low availability of amino acid PET. Another factor preventing wider use of amino acid PET is that it requires experienced users for an objective and comparable diagnosis. In this regard, our approach could play an important role because it provides, for the first time, to our knowledge, an objective and easy-to-use way to volumetrically evaluate amino acid PET data from brain tumor patients. We are confident that the availability of the method to the public will further promote amino acid PET internationally and emphasize its value for clinical decision-making.
CONCLUSION
Our deep learning–based 18F-FET PET segmentation allows a reliable, robust, and fully automated evaluation of MTV in patients with brain tumors. The method alleviates the need for extensive image preprocessing, and its potential for an automated response assessment in patients with gliomas has been demonstrated, fostering translation of volumetric amino acid PET evaluation to clinical routine.
DISCLOSURE
This work was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation; projects 428090865/SPP 2177 [Robin Gutsche, Norbert Galldiks, and Philipp Lohmann] and 491111487). Part of this work was funded by Helmholtz Imaging, a platform of the Helmholtz “Incubator on Information and Data Science.” Norbert Galldiks and Philipp Lohmann received honoraria for lectures from Blue Earth Diagnostics. Norbert Galldiks received honoraria for advisory board participation from Telix Pharmaceuticals. No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: In patients with gliomas, can a fully automated response assessment based on amino acid PET achieve results similar to those of an expert?
PERTINENT FINDINGS: A deep learning–based tumor detection and segmentation tool based on 699 18F-FET PET scans from 555 patients with brain tumors showed high accuracy for lesion detection and segmentation. Further, changes in MTV as evaluated and outlined by the automated segmentation tool were a significant determinant of disease-free and overall survival, in agreement with manual assessment by an expert.
IMPLICATIONS FOR PATIENT CARE: The tumor detection and segmentation tool allows for a fully automated, easy-to-use, objective brain tumor diagnosis and response assessment based on amino acid PET and has the potential to be an important building block to further promote amino acid PET and to strengthen its clinical value.
ACKNOWLEDGMENTS
We thank Silke Frensch, Suzanne Schaden, Trude Plum, Natalie Judov, Kornelia Frey, and Lutz Tellmann for assistance with the patient studies, and we thank Johannes Ermert, Silke Grafmüller, Erika Wabbals, and Sascha Rehbein for radiosynthesis of 18F-FET.
Footnotes
Published online Aug. 10, 2023.
- © 2023 by the Society of Nuclear Medicine and Molecular Imaging.
REFERENCES
- Received for publication March 14, 2023.
- Revision received May 31, 2023.