Visual Abstract
Abstract
Radiomics has been applied to predict recurrence in several disease sites, but current approaches are typically restricted to analyzing tumor features, neglecting nontumor information in the rest of the body. The purpose of this work was to develop and validate a model incorporating nontumor radiomics, including whole-body features, to predict treatment outcomes in patients with previously untreated locoregionally advanced cervical cancer. Methods: We analyzed 127 cervical cancer patients treated definitively with chemoradiotherapy and intracavitary brachytherapy. All patients underwent pretreatment whole-body 18F-FDG PET/CT. To quantify effects due to the tumor itself, the gross tumor volume (GTV) was directly contoured on the PET/CT image. Meanwhile, to quantify effects arising from the rest of the body, the planning target volume (PTV) was deformably registered from each planning CT to the PET/CT scan, and a semiautomated approach combining seed-growing and manual contour review generated whole-body muscle, bone, and fat segmentations on each PET/CT image. A total of 965 radiomic features were extracted for GTV, PTV, muscle, bone, and fat. Ninety-five patients were used to train a Cox model of disease recurrence including both radiomic and clinical features (age, stage, tumor grade, histology, and baseline complete blood cell counts), using bagging and split-sample-validation for feature reduction and model selection. To further avoid overfitting, the resulting models were tested for generalization on the remaining 32 patients, by calculating a risk score based on Cox regression and evaluating the c-index (c-index > 0.5 indicates predictive power). Results: Optimal performance was seen in a Cox model including 1 clinical biomarker (whether or not a tumor was stage III–IVA), 2 GTV radiomic biomarkers (PET gray-level size-zone matrix small area low gray level emphasis and zone entropy), 1 PTV radiomic biomarker (major axis length), and 1 whole-body radiomic biomarker (CT bone root mean square). In particular, stratification into high- and low-risk groups, based on the linear risk score from this Cox model, resulted in a hazard ratio of 0.019 (95% CI, 0.004, 0.082), an improvement over stratification based on clinical stage alone, which had a hazard ratio of 0.36 (95% CI, 0.16, 0.83). Conclusion: Incorporating nontumor radiomic biomarkers can improve the performance of prognostic models compared with using only clinical and tumor radiomic biomarkers. Future work should look to further test these models in larger, multiinstitutional cohorts.
Radiomics is the application of machine learning methods to extract clinically useful information from medical imaging datasets, with an emphasis on systematic, high-throughput mining of “big data” (1–4). Radiomics classifiers have been previously found to enhance prognostic modeling for lung (5,6) and head and neck (7,8) cancers. Recently, this approach has also been applied to cervical cancer, where it has been observed that various radiomic features of the tumor periphery and vascular invasion from PET/MRI are prognostic for locoregional recurrence (9–17). However, PET/MRI machines are not always accessible, particularly in underresourced settings, compared with more established PET/CT techniques (18).
In addition, radiomic analyses in oncology have investigated the predictive information encoded in tumor features, whereas nontumor features, particularly those related to whole-body structures such as bone, bone marrow, fat, muscle, and other organs, have been less studied, although it is worth mentioning a notable recent exception (19), where PET bone marrow features predicted treatment outcome in locally advanced cervical cancer. More generally, such whole-body features may be associated with immune system function, thereby influencing cancer outcomes (20,21). For example, sarcopenia is associated with the release of inflammatory cytokines, such as tumor necrosis factor and interleukin-6 (22), and is a putative marker of disease severity and predictor of outcomes in women with cancer (23,24). Along the same lines, obesity and inflamed adipose tissue are known to impact systemic inflammatory markers and alter the tumor microenvironment (25). Such observations indicate the potential of whole-body imaging, and more generally nontumor features, to provide additional prognostic information beyond that available from tumor features alone. Whole-body PET/CT, with its relatively low cost and widespread availability, is an example of an imaging modality that could provide such information in a cost-effective manner.
Presently, risk-stratification in cervical cancer predominantly depends on clinical examination and standard imaging evaluations. Treatment failures are common, particularly in patients with locoregionally advanced disease, where rates of disease progression may be 30% or more (26). Improved methods to risk-stratify patients with cervical cancer are needed, to appropriately select patients for intensive treatment approaches and identify patients who may selectively benefit from novel therapeutic strategies, such as immunotherapy (27). We therefore sought to determine whether nontumor radiomic features associated with treatment outcomes could be identified in cervical cancer patients undergoing treatment with chemoradiotherapy and imaged using whole-body PET/CT.
MATERIALS AND METHODS
Study Design, Population, and Sampling Methods
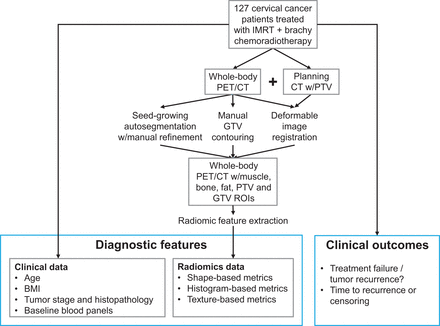
The University of California San Diego institutional review board approved this retrospective cohort study and the requirement to obtain informed consent was waived. We initially identified 245 patients with newly diagnosed, previously untreated, biopsy-proven locoregionally advanced (stage IB–IVA) carcinoma of the cervix treated with chemoradiotherapy at our institution between April 2006 and September 2019. We included patients who underwent pretreatment 18F-FDG PET/CT (PET/CT) and treatment with intensity-modulated radiation therapy (IMRT) followed by intracavitary brachytherapy, resulting in a final cohort of 127 patients (Fig. 1).
Workflow for patient sampling, segmentation, and extraction of high-throughput radiomic features for downstream analysis. BMI = body mass index; Brachy = Brachytherapy; IMRT = intensity-modulated radiation therapy; PET/CT = 18F-FDG PET/CT; ROI: region of interest.
The 127-patient cohort was divided into a training set of 95 patients (75%) and a test set of 32 patients (25%), with 23 and 7 events, respectively. The choice of a 75–25 split was made on the basis of a desire to maintain a sufficient number of events in the training set to be able to adequately train radiomics-based predictors while still keeping enough events in the test set to validate the models. It is important to note that this choice is somewhat arbitrary and, therefore, any conclusions made from model training and validation must be carefully assessed for robustness using extensive bootstrap resampling methods.
Model training and validation consisted of 4 steps region-of-interest (ROI) definition and feature extraction in the entire cohort, identification of robust features in the training set, forward stepwise feature selection from the subset of robust features, and model validation based on comparison of c-indices in the training set and test set. The primary outcome was time from diagnosis to first instance of locoregional or distant cancer recurrence, or censoring, whichever occurred first.
PET/CT Imaging Methods
The pretreatment PET/CT images were acquired on analog Discovery (GE Healthcare) machines, with CT images constructed using filtered backprojection reconstruction for 512 × 512 × 1 voxels and PET images constructed with 1 of 2 settings: ordered-subset expectation maximization (OSEM) reconstruction, with 20 subsets and 2 iterations, using a 4.0-mm gaussian filter cutoff, a 128 × 128 matrix, and a lutetium-yttrium oxyorthosilicate (LYSO) crystal; or OSEM reconstruction with time-of-flight measurements and point-spread function modeling (VUE Point FX; GE Healthcare), with 24 subsets and 2 iterations, using a 5.0-mm gaussian filter cutoff, a 192 × 192 matrix, a 9 × 6 LYSO crystal, and Sharp iterative reconstruction quantitation.
ROI Definitions
We generated ROI segmentations using the workflow illustrated in Figure 1. Three clinical experts manually contoured the gross tumor volume (GTV) for each patient based on the presence of focal hypermetabolic activity within the cervix as well as CT-based anatomic evidence of the primary mass lesion. Grossly involved lymph nodes were not included in the GTV for this study. Additionally, a planning target volume (PTV) consisting of the gross tumor, cervix, uterus, parametria, and pelvic lymph nodes, with a 5- to 15-mm planning margin, were defined on the simulation CT scan by the treating radiation oncologist (5 different radiation oncologists in total), then registered to the whole-body PET/CT using deformable image registration implemented in MIM Maestro (MIM Software Inc.) (28).
For whole-body ROI segmentations, we used a semiautomated approach combining seed-growing (29) and manual editing for muscle contours (Fig. 2). Seed-growing settings were defined as follows: bone – lower bound: 100 Hounsfield units (HU), upper bound: 1,129 HU, tendril diameter: 1 cm, filling level: none; fat – lower bound: −157 HU, upper bound: −123 HU, tendril diameter: 3 cm, filling level: medium; muscle – lower bound: −23 HU, upper bound: 142 HU, tendril diameter: 3 cm, filling level: medium. Additionally, to differentiate skeletal muscle from other smooth or cardiac muscle, further manual editing of the muscle contour was performed. To do this, an exclusion region was generated consisting of internal organs interior to the rib cage and body wall, extending craniocaudally from the trachea to the vagina, and laterally to encompass the breasts (and implants, if present), lungs, and mediastinal contents at the thoracic level, the abdominal organs at the abdominal level, and the reproductive organs at the pelvic level. The psoas muscles were included in the muscle volume rather than in the exclusion region.
Sample output from autosegmentation of whole-body bone, fat, and muscle contours (with manual muscle contour refinement).
Feature Extraction
Radiomics features were extracted using the PyRadiomics (30) software package (version 3.0). For each of the 5 structures (GTV, PTV, bones, muscle, and fat), we calculated all nonredundant features available in PyRadiomics. PyRadiomics can calculate up to 111 features for a given contour and image (17 shape, 19 first-order, 24 gray level correlation matrix [GLCM], 16 gray level run-length matrix [GLRLM], 16 gray level size-zone matrix [GLSZM], 14 gray level difference matrix [GLDM], 5 neighboring gray tone difference matrix [NGTDM]). Of these, 4 shape features can be excluded for redundancy reasons: voxel volume, which is just an approximation of mesh volume, and compactness 1, compactness 2 and spheric disproportion, all of which are completely determined by sphericity. Two first-order features can similarly be ignored: total energy, which is completely determined by the energy and the mesh volume, and the SD, which is just the square root of the variance. Finally, 2 GLCM features, dissimilarity and homogeneity 2, are likewise deprecated as they are equal to the difference average and inverse difference moment, respectively.
These redundancy restrictions result in 193 total features per structure, including 13 shape features and 90 each of CT-based and PET-based intensity features (17 first-order, 22 GLCM, 16 GLRLM, 16 GLSZM, 14 GLDM, 5 NGTDM), resulting in 965 radiomic features. All CT images were resampled, using B-spline interpolation (the default for PyRadiomics), to a 0.98 × 0.98 × 2.5 mm resolution, and all PET images were resampled to a 5.47 × 5.47 × 3.27 mm resolution, based on the lowest resolutions of PET and CT images in the dataset. Radiomic features were extracted using a 25 HU fixed bin width for CT and a 0.5 SUV fixed bin width for PET, based on recommendations from previous radiomic studies (31–34). Notably, choosing the highest resolution PET and CT dimensions results in anisotropic voxels, which previous studies have shown can influence some features, particularly those related to texture matrices (35). Future work systematically assessing how results change with voxel geometry, as well as with use of alternative spline interpolation schemes, is merited. Nevertheless, to the extent that using a consistent choice of settings identifies robust predictors, any findings from this study remain valuable and accessible to the broader community, as long as users make sure to exactly reproduce the extraction settings that are used here.
We also extracted 9 baseline clinical features: age at diagnosis (y), body mass index (kg/m2), tumor histopathology (adenocarcinoma vs. squamous cell carcinoma), stage (I-II vs. III-IVA), and baseline complete blood counts (white blood cells [k/μL], neutrophils [k/μL], hemoglobin [g/dL], and platelets [k/μL]), resulting in 974 candidate features for prediction.
Feature Reduction
To prevent model overfitting and numeric instability due to noise, we first sought to confine our initial set of 974 features to a subset of features that were significantly associated with recurrence. Feature reduction was accomplished through a “bagging” procedure (36). Each bag consisted of a random subset of 57 patients from the training set, with the “out-of-bag” sample consisting of the remaining 38 patients from the training set. For each of the 974 features, we used the bagged subset to train a univariate Cox proportional hazards model, with a single regression coefficient. This process was repeated for 1,000 different bagging subsets, resulting in a 974 × 1,000 matrix of feature coefficients. From this matrix, 99% CIs for each of the 974 coefficients were computed. A feature was defined as robust if the 99% CI excluded zero, indicating a statistically significant association with outcome, for a P value cutoff of 0.01. This process resulted in a final set of robust features (hereafter, candidate biomarkers) from the initial feature set. A table of univariate hazard ratios and CIs resulting from this procedure is shown in the Supplemental Table 1 (supplemental materials are available at http://jnm.snmjournals.org), for all the clinical features as well as for all robust radiomic biomarkers.
Forward Stepwise Feature Selection and Final Model Selection and Validation
We next sought to identify an optimal subset of candidate biomarkers to include as variables in a final Cox model. Potential model variables were selected from the candidate biomarkers using forward stepwise selection. We ran 100 split-sample validations for a model that included a candidate biomarker that we were considering adding to our model. We generated the corresponding 95% CI for each of the model coefficients, as well as the corresponding average out-of-bag c-indices. Additional biomarkers were included in the final model, if they increased the out-of-bag c-index while also maintaining model stability (i.e., the 95% CI for the coefficient estimates for all the model covariates excluded 0). This process was iteratively repeated until either the c-index peaked or there were no more biomarkers that could be stably added. To isolate the effects of tumor radiomic features, and thereby assess the added value of nontumor radiomic information, we repeated the stepwise selection considering only clinical and GTV radiomic biomarkers.
Lastly, a Cox model was trained on the entire (n = 95) training set using the subset of optimal biomarkers selected on each round of forward stepwise selection and evaluated on the unseen test set (n = 32) to test for generalization and overfitting, based on the c-index. To estimate the uncertainty of the c-index, we generated 100 bootstrap resamples each on both the training and the test sets. The likelihood ratio for goodness-of-fit while accounting for Bonferroni multiple-hypothesis-testing corrections as well as the proportional hazards assumption were both tested using the package “survival” (version 3.2-3) in R (R Core Team and R Project for Statistical Computing; www.r-project.org) (37). All P values were 2-sided unless otherwise indicated.
RESULTS
Sample characteristics are given in Table 1. The median follow-up time was 2.12 y in the training set and 2.42 y in the test set.
Sample Characteristics for Training and Test Sets
Following our feature reduction process, the final feature set included 42 candidate biomarkers: 1 clinical biomarker (stage III-IVA vs. I-II), 9 PTV radiomic biomarkers (6 shape, 1 CT-based, 2 PET-based), 13 GTV radiomic biomarkers (1 shape, 1 CT-based, 11 PET-based), 5 muscle radiomic biomarkers (3 shape, 2 CT-based), 10 bone radiomic biomarkers (8 CT-based, 2 PET-based), and 4 fat radiomic biomarkers (1 shape, 3 CT-based). Supplemental Figure 1 shows a hierarchical clustering dendrogram and labeled heat map of these biomarkers, based on the cosine similarity distance, indicating a high degree of collinearity and redundancy.
The forward stepwise selection process resulted in an out-of-bag c-index that peaked after 5 rounds of feature addition, as shown in Figure 3. Furthermore, comparison with the corresponding stepwise selection using only the stage and GTV radiomic biomarkers demonstrated that the prognostic value from nontumor radiomics was additive to tumor radiomics, as this reduced stepwise selection peaked after only 3 rounds and at each round had an out-of-bag c-index consistently lower than the corresponding c-index that could be obtained when including all biomarkers.
Bootstrap averaged out-of-bag (OOB) c-indices during model training, as a function of the number of biomarkers successively added in the forward stepwise selection. Data are shown for selection on only nonradiomic biomarkers (there is only 1, stage, so the stepwise selection trivially stops after 1 round), selection on stage and GTV radiomic biomarkers, and selection on all biomarkers. As the pool of biomarkers available for selection increases, the average OOB c-index at each round likewise increases, and the stepwise selection takes more rounds to reach an optimum. These results suggest that tumor radiomic information, as encoded in the GTV features, adds predictive power beyond that available with just TNM staging, and that off-tumor radiomic information, as encoded in all the other non-GTV features, further adds predictive power beyond that.
As shown in Figure 4, bootstrap resampling to estimate the c-index distributions indicated that the 95% CIs for both the training- and the test-set c-indices overlapped at all rounds of stepwise selection. However, it is imperative to note that the CIs, particularly for the test set, are quite substantial due to the small number of events.
95% CIs for c-index estimates, on both training and test set data, as a function of the number of features successively added in the forward stepwise selection. Results are shown for stepwise selection with stage only (A), stage and GTV biomarkers (B), and all biomarkers (C). The c-index distributions, from which the 95% CI can be determined, are calculated by bootstrap resampling of both the train and the test data. Circular points indicate median values, and color coded bounds indicate upper and lower CI limits, with green corresponding to results for the training set and red corresponding to results for the test set.
The resulting 5 biomarkers in the model are a categoric biomarker that is 0 if the disease is stage I-II and 1 if it is stage III-IVA (mean: 0.32, SD: 0.47); PTV Major Axis Length (mean: 206 mm, SD: 46 mm); CT bone root mean square (mean: 1,394.5 HU, SD: 97.5 HU); PET GTV GLSZM Small Area Low gray Level Emphasis, hereafter SALGLE (mean: 0.045, SD: 0.046); and PET GTV GLSZM Zone Entropy (mean: 5.16, SD: 1.09). Reassuringly, when the stepwise selection is repeated considering only the subset of stage and GTV biomarkers, the resulting 3 biomarkers that emerge are the same categoric stage biomarker and 2 PET GTV GLSZM biomarkers that are found in the full stepwise selection. Model estimates are displayed in Table 2. The likelihood ratio for the Cox model using all 5 biomarkers is 1.0E−05, whereas for the Cox model using only stage and the 2 GTV biomarkers it is 3.8E−04, both indicating statistically significant goodness-of-fit even when accounting for Bonferroni multiple-hypothesis-testing corrections. All variables satisfied the proportional hazards assumption (P > 0.01), and a 1-way ANOVA test (Supplemental Table 2) indicated that none of the selected biomarkers have a statistically significant dependence on acquisition mode or interrater PTV and GTV segmentation variability (P > 0.05).
Cox Model Hazard Ratio Estimates
The radiomics-based Cox model demonstrates great potential for prognosis and risk stratification, as demonstrated by the results shown in Figures 5 and 6. The predicted receiver-operating-characteristic curve for 2-y cancer recurrence, for Cox models both with and without nontumor radiomic biomarkers, lies above the diagonal, and the Kaplan–Meier curves stratifying patients into high- and low-risk groups yields improved hazard ratio estimates compared with stratification based on early- and late-stage groups.
Two-year receiver-operating-characteristic curve for model based on stage only, stage + GTV radiomic biomarkers, and all biomarkers, evaluated on the entire 127-patient cohort. Solid black line corresponds to an area under the curve of 0.5, indicating no predictive performance.
(A) Kaplan–Meier curve based on stratification into early- and late-stage groups. We also list a hazard ratio (HR), with 95% CI and logrank P value. (B) Corresponding Kaplan–Meier curve showing stratification into high- and low-risk groups, based on a stage + GTV radiomic risk score cutoff. The cutoff that minimizes the P value, by coincidence, happens to be at a value such that the number of data points in the high- and low-risk groups equals the corresponding number of late- and early-stage cases. (C) Corresponding Kaplan–Meier curve based on the risk score that fully incorporates stage, GTV radiomic, and non-GTV radiomic biomarkers. The cutoff is chosen so that the number of data points in the high- and low-risk groups equals the corresponding number of late- and early-stage cases. (D) Kaplan–Meier stratification with the model in C but with the cutoff chosen to minimize the P value.
DISCUSSION
Our results suggest that a radiomics model incorporating nontumor radiomic biomarkers leads to improved prognostic modeling of cancer recurrence, compared with using clinical and tumor radiomic biomarkers alone. A novel aspect of our study is the inclusion of semiautomatable whole-body radiomic features as candidate biomarkers for outcome prediction. In addition, whereas much work has been done in identifying CT radiomic biomarkers, the incorporation of PET radiomics remains relatively challenging (38–40), due in part to issues related to feature reproducibility (41) and optimal feature selection in the presence of highly redundant features (42). The approach we developed here has identified 2 PET-based biomarkers of the GTV that seem to be robustly correlated to outcome.
Limitations of this study include the single-institution data source and the size of our cohort, with relatively few total recurrence events. Despite our extensive efforts to maintain quality control (Supplemental Table 3 shows the calculated radiomics quality score (2) for this study) and to cull spurious radiomic features, given the persistent possibility of model overfitting, future work to assess the prognostic power of our results on a larger multiinstitutional cohort is needed to validate the particular predictive model developed in this study. Second, to confine the initial candidate feature set, we focused on particular whole-body features related to bone, fat, and muscle, reasoning that these metrics could reflect variation in patients’ global inflammatory state. Further analysis to study radiomic features of other organs, including other reticuloendothelial organs (liver, spleen) would be of interest. Further extensions to this study could include augmenting radiomic information with additional molecular-level details, as in radiogenomics (43,44), or more detailed examination of the 3D spatial dose distribution (45).
The radiomic predictors we have identified in this work all demonstrate relationships to clinically interpretable physiologic information as identified in previous studies. The PTV Major Axis Length is probably the easiest to understand, being approximately a higher resolution version of tumor stage. A 1-way ANOVA test found that early- and late-stage patients had different mean values of this metric with a P value < 1E−05.
We found 2 PET-based metrics, namely the GTV GLSZM SALGLE, which measures the abundance of small-volume, low-activity “patches,” and the GTV GLSZM zone entropy (46), a measure of textural heterogeneity known to be predictive of outcome. Interestingly, these 2 metrics have a significant, but not perfect, negative correlation (ρ = −0.76). This strong association suggests that the SALGLE combined with the zone entropy capture certain specific aspects of metabolic heterogeneity in and around the tumor microenvironment that are most directly predictive of outcome. Encouragingly, these textural metrics are also similar to a class of PET/MRI radiomic biomarkers that were recently identified (15) and externally validated (16) by Lucia et al. More broadly, this finding is consistent with the established result, both in radiomics and in the oncology community more generally, that metabolic heterogeneity is predictive of cancer recurrence (10,47–49).
The final predictor, the CT Bone Root Mean Square, is probably the most novel one. We found that although age and skeletal muscle volume, as candidate prognostic factors, did not withstand our feature selection algorithm, both were strongly associated with the key bone radiomic metric that did come through (CT Bone Root Mean Square). This suggests that the Root Mean Square CT Bone Number is associated with age-related degeneration of skeletal muscle, as occurs in sarcopenia, a known correlate with inflammation (50,51), which in turn has been shown to be predictive of outcome in locally advanced cervical cancer when combined with PET metrics (19). In fact, multiple studies have found that bone and muscle undergo endocrine crosstalk (52–54), leading some to even suggest that sarcopenia in skeletal muscle and osteoporosis in bones might just be 2 sides of the same underlying condition (55).
CONCLUSION
In summary, we found that incorporating radiomic features, including both tumor and nontumor metrics, improved prognostic modeling of disease recurrence in cervical cancer patients compared with using clinical information alone.
KEY POINTS
QUESTION: Can radiomic models that incorporate pretreatment nontumor PET/CT features improve the prognosis of treatment failure in locoregionally advanced cervical cancer patients, compared with models that use only clinical variables or tumor radiomic features?
PERTINENT FINDINGS: In a retrospective analysis of a single-institutional cohort of 127 patients, optimal performance was seen in a Cox model including 1 clinical staging biomarker, 1 shape feature of the PTV, 2 PET-based features of the GTV, and 1 CT-based feature of whole-body bone segmentation. Stratification into high- and low-risk groups, based on the linear risk score from this Cox model, resulted in a statistically significant improvement in the hazard ratio relative to stratification based on clinical stage alone.
IMPLICATIONS FOR PATIENT CARE: These findings indicate that incorporating nontumor PET/CT radiomic information can improve prognosis of cervical cancer patients undergoing standard-of-care treatment and also identify patients who may benefit from alternative or more intensive treatment regimens.
Footnotes
Published online Oct. 28, 2021.
- © 2022 by the Society of Nuclear Medicine and Molecular Imaging.
REFERENCES
- Received for publication May 21, 2021.
- Revision received October 20, 2021.