Visual Abstract
Abstract
Standardized staging and quantitative reporting are necessary to demonstrate the association of 18F-DCFPyL PET/CT imaging with clinical outcome. This work introduces an automated platform, aPROMISE, to implement and extend the Prostate Cancer Molecular Imaging Standardized Evaluation (PROMISE) criteria. The objective is to validate the performance of aPROMISE in staging and quantifying disease burden in patients with prostate cancer who undergo prostate-specific antigen (PSMA) imaging. Methods: This was a retrospective analysis of 109 veterans with intermediate- or high-risk prostate cancer who underwent PSMA imaging. To validate the performance of aPROMISE, 2 independent nuclear medicine physicians conducted aPROMISE-assisted reads, resulting in standardized reports that quantify individual lesions and stage the patients. Patients were staged as having local disease only (miN0M0), regional lymph node disease only (miN1M0), metastatic disease only (miN0M1), or both regional and distant metastatic disease (miN1M1). The staging obtained from aPROMISE-assisted reads was compared with the staging by conventional imaging. Cohen pairwise κ-agreement was used to evaluate interreader variability. Correlation coefficients and intraclass correlation coefficients were used to evaluate the interreader variability of the quantitative assessment (molecular imaging PSMA [miPSMA] index) at each stage. Kendall tau and t testing were used to evaluate the association of miPSMA index with prostate-specific antigen and Gleason score. Results: All PSMA images of 109 veterans met the DICOM conformity and the requirements for the aPROMISE analysis. Both independent aPROMISE-assisted analyses demonstrated significant upstaging in patients with localized (23%, n = 20/87) and regional (25%, n = 2/8) tumor burden. However, a significant number of patients with bone metastases identified on conventional imaging (18F-NaF PET/CT) were downstaged (29%, n = 4/14). The comparison of the 2 independent aPROMISE-assisted reads demonstrated a high κ-agreement: 0.82 for miN0M0, 0.90 for miN1M0, and 0.77 for miN0M1. The Spearman correlation of quantitative miPSMA index was 0.93, 0.96, and 0.97, respectively. As a continuous variable, miPSMA index in the prostate was associated with risk groups defined by prostate-specific antigen and Gleason score. Conclusion: We demonstrated the consistency of the aPROMISE platform between readers and observed substantial upstaging in PSMA imaging compared with conventional imaging. aPROMISE may contribute to broader standardization of PSMA imaging assessment and to its clinical utility in the management of prostate cancer patients.
Prostate cancer is the most common solid tumor in men, with an incidence of nearly 192,000 cases and nearly 30,000 deaths in the United States annually. Accurate staging of a patient with prostate cancer is critical for selection of appropriate treatment strategies, especially as applied to differentiating between those with localized or regional disease who can be treated with curative intent and those with metastatic disease. Whether surgery, radiation, and systemic hormone therapy or chemotherapy are appropriate for a given patient is driven largely by the clinical stage (1). According to the recently updated National Comprehensive Cancer Network guidelines, 99mTc-phosphonate bone scintigraphy (bone scanning) and CT or MRI remain the standard imaging modalities for prostate cancer staging. However, bone and CT scans have demonstrated limited diagnostic accuracy in earlier disease settings (2,3), in turn limiting the accurate staging necessary for optimal prostate cancer management.
Accurate detection of metastatic disease is a particularly important goal because metastatic prostate cancer requires a different treatment approach and carries a significantly worse prognosis than local disease. PET is a noninvasive technique that can image bone and soft tissue in a single modality, evaluate high-grade tumors that may not produce prostate-specific antigen (PSA), and provide quantifiable data using the SUV. However, in prostate cancer, PET tracers that image metabolic pathways, such as 11C-choline, 11C-acetate, and 18F-FDG, suffer from suboptimal sensitivity and specificity in the detection of regional and distant metastatic disease. Recently, small ligands for PET imaging have been developed that target the cell-surface protein prostate-specific membrane antigen (PSMA), which is overexpressed in prostate cancer cells but is also expressed to some extent in other organs and blood vessels (4). Radiopharmaceuticals based on PSMA ligands have demonstrated high diagnostic accuracy for the detection of both regional and distant metastatic prostate cancer. The proPSMA trial demonstrated that PSMA PET/CT has greater staging accuracy than conventional imaging consisting of bone scanning and CT for initial staging of patients with high-risk prostate cancer (5). This supports the use of a single PSMA PET/CT scan rather than 2 conventional imaging modalities in this setting.
Recent efforts in standardizing the assessment of PSMA scans have resulted in several PSMA PET evaluation and reporting systems, including the PSMA Reporting and Data System, the system of the European Association of Nuclear Medicine, and the Prostate Cancer Molecular Imaging Standardized Evaluation (PROMISE) (6–8). Although all the proposed criteria are focused on the characterization of individual PSMA lesions based on the location and on the definition of significant uptake, the PROMISE standard is also proposing a patient-level staging (molecular imaging TNM), which is based on the detection and location of the disease in the PSMA PET/CT image. A recent study comparing such standardized assessments has shown that they have a high interreader reproducibility (9,10).
However, the adoption of PROMISE criteria in routine clinical practice and investigational studies is limited by the fact that it must be done manually and is labor-intensive. The manual work can be greatly facilitated through automation by deep-learning image analysis. The structural radiologic processes, including the segmentation of anatomic structures (from CT), can be automated to contextualize and characterize the functional imaging. The application of deep learning in automating the whole-body segmentation in PET/CT is the foundational framework for automating the PROMISE criteria. In this study, we introduce and evaluate the analysis of PSMA PET images through aPROMISE, a deep-learning platform to both automate standardized staging and generate a fully quantitative assessment of PSMA-defined disease burden at the lesion and patient levels.
MATERIALS AND METHODS
Patient Population
The purpose of our study was to evaluate the performance of the aPROMISE technology in standardizing the staging and quantification of prostate cancer. This investigation was a retrospective analysis of 109 veterans with unfavorable intermediate- and high-risk primary prostate cancer who underwent 18F-DCFPyL PET/CT under clinical trial NCT03852654, a single-arm trial of PSMA PET/CT on veterans who also underwent conventional imaging with bone scanning, CT, or MRI. The study was approved by the local institutional review board at a Veterans Affairs hospital (PCC 2018-100989), with a waiver of individual informed consent.
Study Design
To validate the performance of aPROMISE, 2 independent board-certified nuclear medicine physicians (3 y of clinical experience) reviewed the PSMA images with the assistance of aPROMISE. No prior instructions were given, and the readers solely and independently relied on the aPROMISE workflow. aPROMISE provides the reader with automated segmentation and quantification of lesions with a preselected molecular imaging TNM type. The reader can choose to accept or override the aPROMISE automated selections at the level of each individual lesion. A final report is autogenerated on the basis of the aPROMISE-assisted read.
First, the aPROMISE-assisted staging was evaluated against conventional-imaging staging obtained from the routine clinical reports. Conventional imaging in every patient included 99mTc-methylene diphosphonate bone scanning or 18F-NaF PET/CT, and CT or MRI of the pelvis. Second, we evaluated the reproducibility of the staging and lesion quantification between the 2 independent aPROMISE-assisted reads. Finally, we evaluated the clinical association of quantitative PSMA uptake (molecular imaging PSMA [miPSMA] index) with 2 baseline clinical variables: Gleason score and PSA value. All patients were staged into 1 of 4 distinct categories: miN0M0 (localized disease and absence of regional lymph node or distant metastatic disease), miN1M0 (regional lymph node disease but absence of distant metastatic disease), miN0M1 (absence of regional lymph node but presence of distant metastatic disease), and miN1M1 (presence of both regional lymph node and distant metastatic disease).
aPROMISE and miPSMA Index
aPROMISE (version 1.1), a class II software as a medical device, is a web application developed by EXINI Diagnostics AB to standardize and quantify PSMA imaging in prostate cancer. aPROMISE is enabled with deep learning that automatically analyzes the CT image to segment anatomic regions in detail, including individual vertebrae, ribs, pelvic bones, and soft-tissue organs such as the prostate (Fig. 1). The anatomic contextualization of the molecular image is used to stage the patient on the basis of the location and extent of the primary tumor in the prostate and of the disease in the local or regional pelvic lymph nodes and in the distant metastases. Subsequently, the PET image is analyzed to detect target lesions. aPROMISE technology enables implementation of standard guidelines such as PROMISE in standardizing PSMA assessment (6). Merging the target lesion information with the anatomic location, the technology quantifies each target lesion in terms of both intensity and volume and summarizes by tissue type to generate the miPSMA index. The aPROMISE report is created automatically, with both aggregated information and detailed information on a per-lesion basis. Manual controls are provided as fallback to augment automatic analysis.
Deep-learning–enabled segmentation of anatomic context in low-dose CT component of PET/CT. Individual color represents respective segmented organ. aPROMISE technology enables automated segmentation of reference organs and anatomic delineation of disease in prostate tumor, regional lymph node, and distant metastases.
In the PROMISE criteria, Eiber et al. (6) defined the miPSMA score of a lesion as 0 when uptake is below the level in the aorta, 1 when uptake is between the levels in the aorta and liver, 2 when uptake is between the levels in the liver and the parotid gland, and 3 when uptake is above the level in the parotid gland. The miPSMA lesion index is a continuous extension of these criteria, defined by linear interpolation from the lesion SUVmean and from the aorta and liver SUV references as follows:


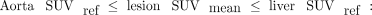

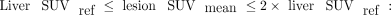
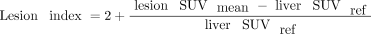
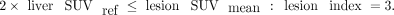
The use of the parotid gland as a threshold has been replaced by 2 times the liver reference since it is not certain that the parotid glands are included in all PSMA PET/CT scans. For each molecular imaging TNM type, lesion uptake is aggregated into the intensity-weighted total lesion uptake volume. This PSMA index is defined as
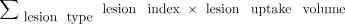 for extent of disease in any lesion type (primary tumor [T stage], local or regional pelvic nodes [N stage], or distant metastases [M stage, which is further denoted as “a” for metastatic lymph nodes, “b” for bone metastases, and “c” for visceral organ metastases]).
for extent of disease in any lesion type (primary tumor [T stage], local or regional pelvic nodes [N stage], or distant metastases [M stage, which is further denoted as “a” for metastatic lymph nodes, “b” for bone metastases, and “c” for visceral organ metastases]).
Statistical Analysis
Descriptive statistics were used to compare the aPROMISE staging with conventional-imaging staging. Cohen pairwise κ-agreement was used to evaluate the interreader variability of aPROMISE-assisted staging (miN0M0, miN1M0, and miN0M1). Spearman and Kendall correlation coefficients were used to evaluate the interreader variability of the quantitative assessment (miPSMA index) of each stage. Student t testing was used to evaluate the miPSMA index values (in tumor) in the risk groups defined by PSA and Gleason score. All statistical analyses were performed using R, version 4.0.2.
RESULTS
The analysis included 109 consecutive patients, whose baseline characteristics are detailed in Table 1. Conventional imaging staged 87 of the 109 patients as having N0M0 disease, 8 patients as having N1M0 disease, 14 patients as having N0M1 disease, and no patients as having N1M1 disease. All 14 of the N0M1 patients were found to have bone metastasis (N0M1b) on conventional staging by 18F-NaF PET/CT and did not undergo 99mTc-methylene diphosphonate bone scanning.
Patient Characteristics (n = 109)
The duration of the aPROMISE-assisted read, from selecting a patient to generating a complete report, was recorded to be a mean of 3.2 min (range, 1.8–5.1 min) per scan for reader 1 and 3.4 min (range, 2.3–5.8 min) for reader 2. The comparative assessment of conventional against aPROMISE-assisted PSMA staging is detailed in Table 2. Both aPROMISE-assisted PSMA analyses demonstrated significant upstaging in patients with localized and regional tumor burden and downstaging in patients who were positive for distant bone metastasis by 18F-NaF PET/CT. In aPROMISE-assisted read 1, of the 87 patients who were determined to be negative for local (N1) or distant (M1) metastatic disease by conventional imaging, 20 (23%) were upstaged in the PSMA imaging assessment to having regional lymph node disease (n = 13) or distant metastatic disease (n = 6). Similarly, of the 8 patients staged as having local pelvic nodal disease only (N1), 2 (25%) were upstaged to having distant metastatic disease also. Notably, a significant population (4/14, 29%) with bone metastatic disease by conventional imaging were downstaged by aPROMISE-assisted PSMA imaging. Examples of downstaged aPROMISE-assisted PSMA reads against 18F-NaF reads are demonstrated in Figure 2. aPROMISE-assisted read 2 had observations similar to those on conventional imaging (Table 2).
aPROMISE-PSMA Staging Reads vs. Local and Distant Metastatic Staging by Conventional Imaging
Example of patients who were negative in aPROMISE-assisted reads of 18F-DCFPyL scans (A and C, axial images) compared with Na18F (B and D, axial images) and were downstaged from N0M1 to N0M0.
Interobserver Reproducibility of aPROMISE Reads
The 2 independent aPROMISE-assisted read are compared in Table 3. The κ-agreement between them was 0.82 for categorization of patients with miN0M0, 0.90 for patient with miN1M0, and 0.77 for patients with miN0M1b. Among all stages, the relatively modest discrepancy in aPROMISE-associated reads was most notable for isolated low-intensity bone lesions. The quantitative reproducibility of miPSMA index in the cases that were categorized the same in the 2 independent aPROMISE-assisted reads—miN0M0 (n = 66), miN1M0 (n = 17), miN0M1(n = 12)—is illustrated in Figure 3. The Spearman correlation was 0.93, 0.96, and 0.97, respectively.
Local and Distant Metastatic Staging by aPROMISE-PSMA Read 1 Against aPROMISE-PSMA Read 2
Quantitative reproducibility of miPSMA index in patients who were categorized the same in 2 independent aPROMISE-assisted reads: miN0M0 (A), miN1M0 (B), and miN0M1(C). In A, 1 patient was excluded because of a manual segmentation error that incorporated bladder. Cor = correlation.
aPROMISE miPSMA Index
As a continuous variable, miPSMA index in the prostate tumor of all patients (n = 109) was correlated with PSA value (t = 0.30; P < 0.0001). Figure 4 shows the miPSMA index values in the prostate, stratified in risk groups defined by PSA and separately by Gleason score. There was a significant difference in values between patients with a PSA of 10 ng/mL or lower (median, 17.61; interquartile range, 8.75–44.63) and patients with a PSA of 20 ng/mL or higher (median, 54.63; interquartile range, 27.55–80.79) (P = 0.05). Similarly, the PSMA index values of prostate tumors with a Gleason score of 3 + 3 (median, 19.45; interquartile range, 9.97–23.54) was significantly lower than that of tumors with a Gleason score of at least 4 + 3 (median, 32.74; interquartile range, 15.38–54.63) (P = 0.01).
miPSMA index values in prostate, stratified by PSA (A and B) and separately by Gleason grade (C).
DISCUSSION
The aPROMISE-assisted independent staging and the quantitative assessments of total disease burden were found to be consistent and reproducible between readers. Integrating PSMA assessment tools into the clinical workflow could allow for automation to provide efficiency, consistency, and accuracy in the staging and quantification of PSMA PET/CT. This study also demonstrated that aPROMISE-assisted reads for PSMA PET/CT detected significantly more regional and metastatic suggestive lesions than were identified by conventional imaging.
The ability of PSMA imaging to detect a greater number of suspected metastatic lesions than can be detected by conventional bone scanning or CT has been evident across multiple studies (11–14). The frequency of upstaging in nodal and distant metastasis by PSMA PET/CT, compared with conventional imaging, in this cohort of patients with intermediate- or high-risk prostate cancer was in line with previous reports. Notably, the biologic dimension of PSMA in evaluating suspected metastatic disease was particularly apparent when comparing findings from 18F-NaF with those from PSMA imaging. Of the 14 patients categorized as M1b through 18F-NaF scans, 4 (29%) were called negative in aPROMISE-assisted reads of their respective PSMA scans. As a bone metabolic scan, 18F-NaF imaging is known to be susceptible to nonpathophysiologic features in bone such as trauma, degenerative changes, and fibrous dysplasia. Of these 4 patients with lesions seen on 18F-NaF imaging but not on PSMA PET/CT, 2 demonstrated lesions that appeared more likely to be benign on PSMA PET/CT but were called positive on the corresponding 18F-NaF imaging. The other 2 patients with discordant findings between 18F-NaF and PSMA imaging had suggestive sclerotic bone lesions that were not seen on the aPROMISE reads because of low PSMA intensity in the lesions. One of these 2 patients underwent curative-intent radical prostatectomy and remains free of biochemical recurrence almost 1 y after surgery, without additional therapy. The other patients delayed treatment and instead underwent a repeat 18F-NaF examination 6 mo later that showed no interval change in the bone lesion but did show progression within soft tissue. In these 2 cases, clinical follow-up was more consistent with the PSMA PET staging than with the 18F-NaF imaging. A more comprehensive comparison of PSMA and 18F-NaF imaging is beyond the scope of this study but will be done in a separate follow-up analysis.
Interreader agreement on the interpretation of PSMA PET/CT has been evaluated mostly using 68Ga-PSMA11 PET/CT. Fendler et al. evaluated interreader agreements in 50 patients with primary disease and after biochemical recurrence and found κ values of 0.62 for primary tumor, 0.74 for nodes, and 0.88 for bone lesions (15). In a more homogeneous biochemically recurrent population consisting of patients with PSA levels of up to 0.6 ng/mL, Miksch et al. demonstrated κ values of 0.76 for primary tumor, 0.73 for nodes, and 0.58 for bone lesions (16). In a study focused exclusively on 50 patients who underwent 18F-DCFPyL PET, an intraclass correlation coefficient of 0.79 was derived for nodal disease (17). Similarly, the manual reproducibility of following the PROMISE classification has been reviewed and reported by Toriihara et al., who demonstrated moderate interreader agreement (0.67) for molecular imaging TNM classification in PSMA PET/CT (9). The agreement between the aPROMISE-assisted reads in our study compares favorably against these prior evaluations (Cohen κ > 0.75), with a notably quick reading time (mean, 3.2 and 3.4 min per scan). One reader in our study had considerably more prior experience in the interpretation of PSMA PET/CT than did the other. Still, a high degree of agreement was noted. The readers in our study did not get any strict guidance on lesion detection, nor did they receive any formal training on the PROMISE criteria. The findings may suggest that an aPROMISE-assisted read that involves automated segmentation, localization, and lesion preselection may nudge readers toward a moderately high agreement irrespective of their prior experience. This hypothesis warrants a multicenter, multireader study for validation.
Quantitative metrics of disease burden may further enhance the prognostic and predictive power of imaging. Currently, the automated bone scan index (aBSI) is the only Food and Drug Administration–cleared software as a medical device that has been prospectively validated in a registration study as a prognostic imaging biomarker for metastatic prostate cancer (18). The STAMPEDE trial investigated the addition of radiation to the primary tumor in M1 patients. In a post hoc analysis that used aBSI to assess disease burden, aBSI was predictive of response to prostate radiotherapy (19). aBSI uses a machine-learning algorithm that preselects and segments the lesions in bone and automatically computes a quantitative total tumor burden in 99mTc planar bone scans (20). In some sense, the miPSMA index for quantification of disease burden defined by PSMA PET/CT can be considered a 3-dimensional analog of aBSI.
However, the automated miPSMA index offers a far more comprehensive assessment of disease burden. The miPSMA index is a continuous extension of the miPSMA score proposed in the PROMISE criteria. Like the miPSMA score, the miPSMA index is the PSMA quantification of an individual lesion in relation to the mean uptake in reference organs. The result, for each lesion, is a linear PSMA-burden quantification that can be summarized by each tissue type (primary tumor [T stage], local or regional pelvic nodes [N stage], or distant metastases [M stage, which is further denoted as “a” for metastatic lymph nodes, “b” for bone metastases, and “c” for visceral organ metastases]). Our study showed an association between miPSMA index in the primary tumor and both Gleason grade and PSA value. This finding is consistent with prior studies reporting that PSMA expression in the primary tumor is associated with a higher Gleason grade and recurrence risk (21,22). We hypothesize that the miPSMA index may be useful for selecting patients for PSMA-targeted radiotherapy, with current trials largely using qualitative assessments of PSMA expression as inclusion criteria. Moreover, there is a potential role for the miPSMA index in conjunction with morphologic findings as a quantitative method of response assessment after treatment.
The purpose of our hypothesis-generating study was to evaluate the performance of the aPROMISE technology for subsequent prospective clinical investigations. The findings here enable future investigations to evaluate any additive benefits of aPROMISE-assisted reads over manual reads of PSMA PET/CT and to assess whether the diagnostic performance of PSMA PET/CT is enhanced when using the aPROMISE software. Our study was limited in the number of independent reads and in its retrospective design. Therefore, the findings and the hypothesis presented here should be validated in a prospectively designed multireader and multiinstitutional study design. In addition, lesions selected by aPROMISE have not been histopathologically validated. However, PSMA PET was shown to have high specificity in several recent studies (23).
Despite these limitations, our study demonstrated the performance of aPROMISE in an independent assessment. Incorporation of aPROMISE and the miPSMA index into subsequent clinical investigations can allow further exploration of the clinical context of their use for prospective validation.
CONCLUSION
aPROMISE-assisted PSMA PET/CT reads generate detailed imaging reports at the whole-patient and lesion levels within minutes. Compared with conventional imaging, aPROMISE assistance upstages patients and reduces interreader variability, even among readers with differing baseline levels of experience. Moreover, aPROMISE-assisted reads may standardize PSMA evaluation. Prospective studies and direct manual comparison studies are required to validate these findings. The miPSMA index is a quantitative measure of lesion volume and relative intensity, is associated with Gleason grade and PSA, and describes overall and tissue-specific tumor burden. Evaluation of the miPSMA index as an imaging biomarker of disease burden is warranted in order to assess prognostic value.
DISCLOSURE
This work was supported by EXINI Diagnostics AB (a wholly owned subsidiary of Progenics Pharmaceuticals Inc.). Nicholas Nickols is a PCF Young Investigator. Johan Brynlofsson and Kerstin Johnsson are employees of EXINI Diagnostics AB. Aseem Anand and Pablo Borelli have received honorary support from EXINI Diagnostics AB. No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: Can the aPROMISE platform generate a consistent and standardized evaluation of PSMA scans?
PERTINENT FINDINGS: The comparison of the 2 independent aPROMISE-assisted reads demonstrated a high κ agreement in staging of patients. As a continuous variable, miPSMA index in the prostate was associated with risk groups defined by PSA values and Gleason scores.
IMPLICATIONS FOR PATIENT CARE: aPROMISE-assisted reads may standardize PSMA evaluation and reduce interreader variability, even among readers with differing baseline levels of experience. The miPSMA index is a quantitative measure of lesion volume and relative intensity, is associated with Gleason grade and PSA, and describes overall and tissue-specific tumor burden.
Footnotes
Published online May 28, 2021.
- © 2022 by the Society of Nuclear Medicine and Molecular Imaging.
REFERENCES
- Received for publication December 23, 2020.
- Revision received April 23, 2021.












