Visual Abstract
Abstract
Artificial intelligence may improve accuracy of myocardial perfusion imaging (MPI) but will likely be implemented as an aid to physician interpretation rather than an autonomous tool. Deep learning (DL) has high standalone diagnostic accuracy for obstructive coronary artery disease (CAD), but its influence on physician interpretation is unknown. We assessed whether access to explainable DL predictions improves physician interpretation of MPI. Methods: We selected a representative cohort of patients who underwent MPI with reference invasive coronary angiography. Obstructive CAD, defined as stenosis ≥50% in the left main artery or ≥70% in other coronary segments, was present in half of the patients. We used an explainable DL model (CAD-DL), which was previously developed in a separate population from different sites. Three physicians interpreted studies first with clinical history, stress, and quantitative perfusion, then with all the data plus the DL results. Diagnostic accuracy was assessed using area under the receiver-operating-characteristic curve (AUC). Results: In total, 240 patients with a median age of 65 y (interquartile range 58–73) were included. The diagnostic accuracy of physician interpretation with CAD-DL (AUC 0.779) was significantly higher than that of physician interpretation without CAD-DL (AUC 0.747, P = 0.003) and stress total perfusion deficit (AUC 0.718, P < 0.001). With matched specificity, CAD-DL had higher sensitivity when operating autonomously compared with readers without DL results (P < 0.001), but not compared with readers interpreting with DL results (P = 0.122). All readers had numerically higher accuracy with CAD-DL, with AUC improvement 0.02–0.05, and interpretation with DL resulted in overall net reclassification improvement of 17.2% (95% CI 9.2%–24.4%, P < 0.001). Conclusion: Explainable DL predictions lead to meaningful improvements in physician interpretation; however, the improvement varied across the readers, reflecting the acceptance of this new technology. This technique could be implemented as an aid to physician diagnosis, improving the diagnostic accuracy of MPI.
Coronary artery disease (CAD) is a major cause of death in the United States (1), making accurate diagnosis critically important. Myocardial perfusion imaging (MPI) is frequently used to diagnose obstructive CAD (2), predict cardiovascular risk (3), or guide treatment decisions (4,5). Artificial intelligence (AI) may be able to improve the diagnostic (6–9) and prognostic accuracy of MPI (10,11); however, it is not feasible or desirable to implement it without physician oversight (12–15). As a result, for the foreseeable future, AI will most likely be implemented as an aid to physician interpretation rather than operating autonomously (12–15).
We recently demonstrated an approach for automated interpretation of MPI by a general-purpose deep-learning (CAD-DL) model, which incorporated 2 methods to explain predictions to physicians (9). These methods for explainability allow physicians to ensure that AI findings are clinically relevant and potentially identify errors in either AI or physician interpretations. Explainable AI is critical to overcoming the “black-box” perception of AI (16,17) and is recognized as an important advancement in a recent AI best practice statement (12). In our initial study, autonomous CAD-DL score had higher diagnostic accuracy for obstructive CAD than expert visual interpretation and quantitative assessment of myocardial perfusion (9). However, it remains to be shown if explainable DL predictions, used as an aid during clinical interpretation, can improve physician interpretation of MPI, which is the likely method for clinical implementation (12).
Accordingly, we performed a prospective study to assess the potential improvement in accuracy of physician interpretation of SPECT MPI that could be achieved by using the explainable DL model as an aid during clinical interpretation, using an external population from sites not used for model training.
MATERIALS AND METHODS
Study Population
We included 240 patients who underwent SPECT MPI with reference coronary angiography within 180 d. Patients were included from 2 sites, Columbia University (n = 125) and Cardiovascular Imaging Technologies LLC (n = 115). No data from these sites were used in the development of CAD-DL. Patients were randomly selected to include a representative patient population with prevalence of obstructive CAD of 50%. Patients underwent imaging with either conventional Anger camera systems (n = 80) or solid-state camera systems (n = 160). Patients underwent stress–rest (n = 158), rest–stress (n = 61) or stress-only (n = 21) imaging protocols with either exercise stress with a symptom-limited Bruce protocol (n = 98) or pharmacologic stress with adenosine (n = 31) or regadenoson (n = 111). Patients underwent either a standard single-isotope (n = 205) or dual-isotope (n = 35) SPECT MPI protocol as previously described (2,18). For comparison, CAD-DL was trained in a population imaged with 58% solid-state camera systems and 42% conventional camera systems, with 69% of patients undergoing pharmacologic stress. The institutional review board at each site approved this study with either signed informed consent or waiver of informed consent.
The study protocol complied with the Declaration of Helsinki and was approved by the institutional review boards at each participating institution. The overall study was approved by the institutional review board at Cedars-Sinai Medical Center. To the extent allowed by data sharing agreements and institutional review board protocols, the data from this article will be shared on written request.
Details regarding invasive coronary angiography and image quantification are available in the supplemental materials (available at http://jnm.snmjournals.org) (19,20).
DL Model Architecture
The architecture for CAD-DL has previously been described in detail (9). In brief, CAD-DL was trained using raw polar maps, preprocessed using Z-normalization (mean of 0 and SD of 1), of myocardial perfusion, wall motion, and thickening (21), as well as age, sex, left ventricular end-systolic volume and end-diastolic volumes, which were all obtained automatically from image data. CAD-DL was trained in a separate population of 3,578 patients, with obstructive CAD present in 63% of patients. CAD-DL was implemented using Python 3.7.3 and Tensorflow 1.14. The training was performed using Titan RTX graphics card (Nvidia). The model was trained using 5-fold cross-validation in the previous study, which did not include any data from the 2 sites in the present study.
CAD-DL includes 2 methods to explain predictions: attention maps and probability maps. Attention maps use gradient-weighted class activation mapping (22) to highlight myocardial regions which contributed most to the prediction. Using these attention maps, each segment from the standard 17-segment American Heart Association model can be assigned to 5 categories to generate the segmental CAD probability map. The CAD probability map marks the degree to which the segments contribute the CAD-DL prediction, to give further insight for the clinical reader.
Physician Interpretation
Three physicians with a range of clinical experience (2 y to >20 y in clinical practice) interpreted all cases in duplicate. Initially, readers interpreted myocardial perfusion images in the conventional manner and were supplied with the following: age, sex, body mass index, past medical history, test indication, electrocardiographic stress response, and images. Readers had access to all image datasets including stress and rest as well as supine and upright when available and gated imaging for all studies (23). Raw data were available for quality control, and standard quantitative measures of function and perfusion were available for all studies. Readers interpreted the overall study with a 5-point scale (normal, probably normal, equivocal, probably abnormal, and definitely abnormal). Readers also interpreted studies with semiquantitative scoring to generate summed stress score (SSS), summed rest score, and summed difference score using the standard 17-segment model.
After the initial interpretation, readers interpreted the study in conjunction with CAD-DL. The clinical prototype of CAD-DL developed previously, with attention and probability maps, was integrated within standard clinical nuclear cardiology software (QPS/QGS Cedars-Sinai Medical Center, Los Angeles, CA). Physicians repeated the interpretation process when informed by CAD-DL results. This process was designed to simulate the use of CAD-DL in clinical practice, where it would be incorporated as an expert second reader. Physicians were trained on how to generate CAD-DL results but were not given specific thresholds for CAD-DL global score, attention map, or CAD-DL probability scores to apply in their new interpretation. No specific instructions were given for adjusting reader interpretations based on CAD-DL results. This was done purposefully so the results would reflect clinical practice whether other factors (e.g., reader confidence in original interpretation, belief in AI interpretation) would influence the degree to which readers alter their interpretation.
Statistical Analysis
In the primary analysis, the diagnostic performance of SSS without DL, SSS with DL, and stress total perfusion deficit (TPD) was evaluated using the analysis and pairwise comparisons of the areas under the receiver-operating-characteristic (ROC) curve (AUC) according to DeLong et al. (24) in order to allow comparisons with automated perfusion assessment. However, in order to more fully assess the impact of DL predictions on reader diagnostic accuracy, we performed an analysis to account for the multiple reader, multiple case (MRMC) design, which accounts for variation related to case variation, reader certainty, and reader skill. In this analysis, a random-effects ROC analysis was used to compare the reader-averaged nonparametric AUC with and without access to DL predictions as previously described (25,26). Additional details are available in the supplemental materials.
RESULTS
Clinical Characteristics and Angiographic Characteristics
In total, 240 patients with a median age of 65 y (IQR 58–73) and 156 (65.0%) male were included in this study. Invasive angiography was performed at a median of 11 d (IQR 3–27 d) after SPECT MPI. Obstructive CAD was present in 120 (50.0%) patients including 11 patients with left main, 84 with left anterior descending coronary artery, 55 with left circumflex coronary artery, and 63 with right coronary artery disease. Characteristics in patients with and without obstructive CAD are shown in Table 1. Characteristics stratified by camera type are shown in Supplemental Table 1. Median age was similar in patients imaged with a conventional or solid-state camera system (median 66 vs. 65, P = 0.858).
Population Characteristics Stratified by Presence of Obstructive CAD
Per-Patient Diagnostic Accuracy
ROC curves for identification of obstructive CAD based on stress perfusion assessment (SSS) are shown in Figure 1. The AUC of physician interpretation with DL (AUC 0.779, 95% CI 0.738–0.850) was significantly higher than that of both physician interpretation without DL (AUC 0.747, 95% CI 0.685–0.809, P = 0.003) and stress TPD (AUC 0.718, 95% CI 0.653–0.782, P < 0.001). The diagnostic accuracy of physician interpretation with DL was similar to CAD-DL operating autonomously (AUC 0.793, 95% CI 0.736–0.849, P = 0.536).
Diagnostic accuracy of stress perfusion for obstructive CAD. Summed stress scores for all readers were averaged to determine reader accuracy with and without deep learning (DL). CAD-DL are results from DL model when operating autonomously.
Diagnostic accuracy for each reader separately is shown in Supplemental Figure 1. There was a trend toward improvement in accuracy for 2 readers (reader 2 AUC 0.750 vs. 0.730, P = 0.115 and Reader 3 AUC 0.751 vs. 0.733, P = 0.068). Reader 1 demonstrated significantly improved accuracy with access to CAD-DL predictions (AUC 0.805 vs. 0.756, P = 0.005). Readers 1 and 2 were less experienced (4 and 2 y, respectively) than Reader 3 (>20 y). For comparison, the AUC of DL operating autonomously was 0.793 (95% CI 0.736–0.849). In the MRMC analysis, reader accuracy was also significantly improved with access to CAD-DL predictions (AUC 0.769 vs. 0.740, P = 0.019).
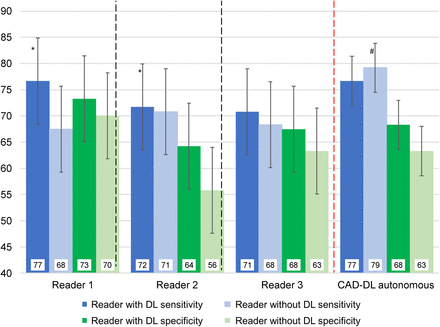
Figure 2 shows reader sensitivity and specificity, using a threshold of SSS > 3, with and without DL. All test characteristics numerically improved when readers had access to explainable DL results, with improvement in both sensitivity and specificity when considering all readers together (both P < 0.01). With matched specificity, CAD-DL had higher sensitivity when operating autonomously than did readers without access to explainable DL results (P < 0.001). However, this difference was not significant when readers had access to DL predictions during interpretation (P = 0.122).
Sensitivity and specificity of reader interpretation with and without deep learning (DL). Summed stress score > 3 was considered abnormal. Two thresholds were established for CAD-DL to match sensitivity of average reader specificity with (dark bars) and without (light bars) access to DL predictions. *P < 0.05 reader with DL compared with reader without DL, #P < 0.001 CAD-DL operating autonomously compared with all readers.
Our study was powered to detect a difference in accuracy for the overall population. However, there was a trend toward improved physician diagnostic performance with DL compared with without DL for both camera systems (solid-state AUC 0.799 vs. 0.774, P = 0.095; conventional AUC 0.740 vs. 0.691, P = 0.014) across imaging protocols (stress–rest AUC 0.775 vs. 0.738, P = 0.009; rest--stress AUC 0.710 vs. 0.690, P = 0.464; and stress-only AUC 0.891 vs. 0.881, P = 0.665), in men (AUC 0.792 vs. 0.772, P = 0.096) and women (AUC 0.714 vs. 0.658, P = 0.028), and in patients undergoing exercise (AUC 0.816 vs. 0.795, P = 0.250) or pharmacologic stress (AUC 0.728 vs. 0.692, P = 0.020). Reader interpretation of ischemia with DL (based on summed difference score) also demonstrated significantly higher AUC than reader interpretation without DL or ischemic TPD (Fig. 3).
Diagnostic accuracy of ischemia for obstructive CAD. Summed difference scores for all readers were averaged to determine reader accuracy with and without deep learning (DL). SDS = summed difference score.
Lastly, reader diagnosis, using a 5-point scale, with DL also demonstrated significantly higher accuracy than reader diagnosis without DL (Supplemental Fig. 2). The reclassification of patients according to reader diagnosis for the 3 readers is shown in Table 2. There was an overall net reclassification improvement of 17.2% (95% CI 9.2%–24.4%, P < 0.001), with improved classification of patients with CAD of 6.1% (95% CI 1.4%–10.3%) and patients without CAD of 11.1% (95% CI 4.8%–16.8%). When interpreting with DL compared to the reader interpreting without DL, there was no difference in the proportion of cases interpreted as equivocal (9.6% vs.8.6%, P = 0.529). Similarly, there was no difference in the proportion of cases with CAD interpreted as definitely abnormal (59% vs. 58%, P = 0.803) or patients without CAD interpreted as definitely normal (26% vs. 23%, P = 0.464).
Net Reclassification of Patients with CAD-DL (Explainable Deep Learning Model) Compared With the Same Readers Without CAD-DL
One case in which all 3 physicians increased their segmental scores in a patient with obstructive CAD is shown in Supplemental Figure 3. One case in which all 3 physicians decreased their segmental scores in a patient without obstructive CAD is shown in Supplemental Figure 4. A description of all cases in which all readers increased or decreased their scores is available in Supplemental Table 2. An example of a case with high CAD-DL score that was not consistently scored as abnormal is shown in Supplemental Figure 5.
Per-Vessel Diagnostic Accuracy
AUCs for identification of obstructive CAD for each vessel are shown in Supplemental Figure 6. Reader diagnostic accuracy with DL (AUC 0.723, 95% CI 0.652–0.793) was significantly better than accuracy without DL (AUC 0.697, 95% CI 0.626–0.768, P = 0.041) for left anterior descending coronary artery disease. Reader interpretation with DL had higher AUC than reader interpretation without DL and stress TPD for left anterior descending disease (P = 0.041 and 0.022, respectively). Reader interpretation with DL was not significantly higher than reader interpretation without DL for left circumflex (AUC 0.727 vs. 0.715, P = 0.529) or right coronary artery disease (AUC 0.776 vs. 0.779, P = 0.597). Reclassification according to vascular territory is shown in Supplemental Table 3.
DISCUSSION
We performed a prospective study in an external population to determine the potential influence of using an explainable DL model as an interpretation aid on physician diagnostic accuracy. We demonstrated that overall physician interpretation significantly improved by using the DL predictions compared with the same physicians interpreting without DL. Additionally, with the aid of DL physician interpretation had higher diagnostic accuracy than quantitative assessment of perfusion. There was a trend toward higher diagnostic performance for every reader, and results were consistent across camera systems, imaging protocols, and patient subgroups. There was some heterogeneity in improvement in physician diagnostic performance; however, there was more uniformity in sensitivity and specificity across readers when interpreting with DL results. All of these advancements were demonstrated despite the relative novelty of the DL tool and lack of physician experience with using the new DL module. Our results suggest that implementing DL as an aid to physician interpretation could significantly improve diagnostic accuracy of MPI.
Several studies have previously demonstrated that AI algorithms can be used to achieve high diagnostic accuracy of SPECT MPI. Arsanjani et al. demonstrated that a support vector machines model improved diagnostic accuracy for obstructive CAD compared with quantitative assessment of perfusion with TPD (27). Betancur et al. demonstrated that a different DL model improved detection of obstructive CAD compared with quantitation of perfusion with TPD on both a regional and a per-patient basis, in a study that included 1,638 patients from 9 centers (6). With matched specificity, DL improved the per-vessel sensitivity to 70% compared with 64% with TPD (P < 0.01) (6). Subsequently our group demonstrated that a DL algorithm using both upright and supine imaging data improved diagnostic accuracy compared with combined TPD analysis (7). Spier et al. demonstrated that a convolutional neural network could classify stress polar maps as normal or abnormal with excellent agreement with expert interpretation (91.1%) (28). More recently we demonstrated that the current model, CAD-DL, had higher diagnostic accuracy for obstructive CAD than physician interpretation or quantitative assessment of perfusion (9). However, all of these studies demonstrated only standalone performance, without physician oversight, which is not practical clinically.
In the current study we addressed an important knowledge gap by determining to what extent access to explainable DL predictions could influence or improve physician interpretation. We demonstrated, in an external population suggesting broader generalizability (12), that overall physician interpretation significantly improved with the aid of CAD-DL compared with the same physicians interpreting without CAD-DL. All readers demonstrated numerically higher AUC with DL, with one reader improving significantly. Importantly, readers had access to the same clinical and imaging information (including quantitative results) during each interpretation, with the only difference being the AI predictions. Overall reader sensitivity and specificity both improved, achieving results similar to CAD-DL operating autonomously. Using CAD-DL as an aid also significantly improved classification of the overall population, patients with CAD, and patients without CAD. Per-vessel diagnostic accuracy was significantly higher for the left anterior descending and similar in other vascular territories. There was also net improvement in classification across all territories. This result was obtained using a patient population separate from the original population used for training, with different population characteristics and prevalence of obstructive CAD suggesting good generalizability of the results. Importantly, the DL model was incorporated into standard clinical interpretation software, which was used by all readers and generated results in <10 s. However, we did not measure the average interpretation time for readers when interpreting with and without the explainable DL predictions, which may be an important consideration for clinical implementation. Overall, our findings suggest that our model could be implemented into clinical practice as an aid to physician interpretation in order to improve the diagnostic accuracy of SPECT MPI.
Although the potential benefits of AI for improving diagnostic accuracy are becoming readily apparent, practical questions about clinical implementation have remained. One step toward clinical implementation is the development of models capable of explaining results to the physician. The CAD-DL model user interface includes 2 methods to explain predictions to the clinicians with the attention and probability maps. In order to replicate future clinical implementation, we instructed physicians on how to access predictions but did not explicitly instruct them on how to incorporate this information. This approach to using DL predictions mirrors future clinical use where factors such as physician experience, confidence in original interpretation, belief in AI, and anchoring bias would influence thresholds for changing interpretation and magnitude of change (29). As a result, and as was seen in our study, the degree to which CAD-DL influences interpretation varies between physicians (Fig. 3). Although there was no clear relationship between reader experience and improvement in accuracy in our study, it is likely that less experienced readers would derive more benefit. One recent study suggested that a deep-learning algorithm could be used by novice physicians to attain interpretation of myocardial perfusion similar to that of experts (30). Additionally, further improvements may be possible if physicians develop more experience with incorporating the DL predictions. Despite this variation, access to DL results led to more uniform sensitivity and specificity across readers. Additionally, as physician experience with DL increases, diagnostic accuracy may improve further. Incorporating AI as an aid to physician interpretation avoids potential medicolegal issues related to using these technologies because the final responsibility for interpretation still lies with the physician.
Our study has a few important limitations. The explainable DL model incorporates several methods of explaining results (attention maps, probability maps, per-vessel probabilities) that are presented simultaneously in the clinical module. We did not elucidate which aspect of the explainable DL predictions led to improved accuracy since they are designed to be interpreted collectively. The per-vessel probabilities convey the likelihood of CAD to physicians while the attention and probability maps direct the physician’s attention to allow them to validate the accuracy of those predictions. Additionally, we did not measure subjective changes in reader confidence but did not identify a change in proportion of studies interpreted as equivocal or definitely normal or abnormal. We used patients from 2 separate sites who underwent a range of stress and imaging protocols. Although this increases the generalizability of our results, it decreases the precision of our estimate regarding the increase in accuracy for any one combination of camera system and imaging protocol. The MRMC analysis accounts for case variation as well as variations in reader certainty, reader skill, and reader response to AI but more precise evaluation of the impact of explainable AI on physician interpretation could be made in a population imaged with a single camera system and imaging protocol. Additionally, we would be able to make more definitive conclusions about the influence of explainable DL results on reader interpretation if additional readers were involved and all readers interpreted a greater number of studies. As will be the case in clinical practice, interpreters in the study had variable exposure and belief in AI models before the study. It is possible that additional experience with using CAD-DL may lead to further improvements in accuracy.
CONCLUSION
We demonstrated that overall physician interpretation significantly improved by using the DL predictions compared with the same physicians interpreting without DL. Implementing DL as an aid to physician interpretation significantly improves diagnostic accuracy of MPI.
DISCLOSURE
This research was supported in part by grant R01HL089765 from the National Heart, Lung, and Blood Institute/National Institutes of Health (NHLBI/NIH) (PI: Piotr Slomka). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Drs. Daniel S. Berman and Piotr J. Slomka and Paul Kavanagh participate in software royalties for QPS software at Cedars-Sinai Medical Center. Dr. Slomka has received research grant support from Siemens Medical Systems. Drs. Berman, Sharmila Dorbala, Andrew J. Einstein, and Edward Miller have served as consultants for GE Healthcare. Dr. Einstein has served as a consultant to W. L. Gore & Associates and has served on a speakers bureau for Ionetix. Dr. Dorbala has served as a consultant to Bracco Diagnostics; her institution has received grant support from Astellas. Dr. Marcelo Di Carli has received research grant support from Spectrum Dynamics and consulting honoraria from Sanofi and GE Healthcare. Dr. Terrence D. Ruddy has received research grant support from GE Healthcare and Advanced Accelerator Applications. Dr. Einstein’s institution has received research support from GE Healthcare, Philips Healthcare, Toshiba America Medical Systems, Roche Medical Systems, and W. L. Gore & Associates. Dr. Keiichiro Kuronuma has received funding support from the Society of Nuclear Medicine and Molecular Imaging Wagner-Torizuka Fellowship grant and the Nihon University School of Medicine Alumni Association Research Grant. No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: Does an explainable DL model, when used as an aid during interpretation, improve physician diagnostic accuracy?
PERTINENT FINDINGS: In this multiple reader, multiple case interpretation study, access to explainable DL results led to meaningful but variable improvements in the accuracy of physician interpretation of MPI. Overall, diagnostic performance improved when physicians had access to DL predictions and readers demonstrated improved classification of patients with and without CAD.
IMPLICATIONS FOR PATIENT CARE: Explainable DL could be implemented as an aid to physician interpretation in order to improve diagnostic accuracy potentially improving patient management and subsequent outcomes.
ACKNOWLEDGMENTS
We thank all the individuals involved in the collection, processing, and analysis of data in this multicenter registry.
Footnotes
Published online May. 5, 2022.
- © 2022 by the Society of Nuclear Medicine and Molecular Imaging.
REFERENCES
- Received for publication December 13, 2021.
- Revision received April 18, 2022.