Abstract
Accurate delineation of the intraprostatic gross tumor volume (GTV) is a prerequisite for treatment approaches in patients with primary prostate cancer (PCa). Prostate-specific membrane antigen PET (PSMA PET) may outperform MRI in GTV detection. However, visual GTV delineation underlies interobserver heterogeneity and is time consuming. The aim of this study was to develop a convolutional neural network (CNN) for automated segmentation of intraprostatic tumor (GTV-CNN) in PSMA PET. Methods: The CNN (3D U-Net) was trained on the 68Ga-PSMA PET images of 152 patients from 2 different institutions, and the training labels were generated manually using a validated technique. The CNN was tested on 2 independent internal (cohort 1: 68Ga-PSMA PET, n = 18 and cohort 2: 18F-PSMA PET, n = 19) and 1 external (cohort 3: 68Ga-PSMA PET, n = 20) test datasets. Accordance between manual contours and GTV-CNN was assessed with the Dice-Sørensen coefficient (DSC). Sensitivity and specificity were calculated for the 2 internal test datasets (cohort 1: n = 18, cohort 2: n = 11) using whole-mount histology. Results: The median DSCs for cohorts 1–3 were 0.84 (range: 0.32–0.95), 0.81 (range: 0.28–0.93), and 0.83 (range: 0.32–0.93), respectively. Sensitivities and specificities for the GTV-CNN were comparable with manual expert contours: 0.98 and 0.76 (cohort 1) and 1 and 0.57 (cohort 2), respectively. Computation time was around 6 s for a standard dataset. Conclusion: The application of a CNN for automated contouring of intraprostatic GTV in 68Ga-PSMA and 18F-PSMA PET images resulted in a high concordance with expert contours and in high sensitivities and specificities in comparison with histology as a reference. This robust, accurate and fast technique may be implemented for treatment concepts in primary prostate cancer. The trained model and the study’s source code are available in an open source repository.
In patients with newly diagnosed prostate cancer (PCa), accurate contouring of the intraprostatic gross tumor volume (GTV) is mandatory for successful fusion-biopsy guidance (1). Additionally, focal therapy approaches such as focal dose escalation in radiotherapy (2) rely on an accurate definition of the intraprostatic GTV.
Prostate-specific membrane antigen PET (PSMA PET) has recently been established for initial staging in primary PCa patients (3). It is also increasingly used to improve intraprostatic lesion detection (4–4), focal therapy guidance (7), and noninvasive PCa characterization (8). Most of the studies evaluated 68Ga-PSMA-11 as the radiopharmaceutical. However, 18F-PSMA-1007 is increasingly used, and Kuten et al. reported that 18F-PSMA-1007 may detect additional low-grade lesions (9). In a recent work, manual and semiautomatic contouring approaches for 68Ga-PSMA PET images were validated (10). Although good results (sensitivity and specificity > 80%) were obtained for most of the contouring approaches, some methodologies showed a rather poor performance (sensitivity and specificity < 70%). This is in line with a Dice-Sørensen coefficient (DSC) varying between 0.56 and 0.8 for the manual contours, which indicates that PSMA PET–based GTV definition underlies a substantial interobserver variability. Actually, no validated contouring technique for 18F-PSMA PET was proposed.
The implementation of an automatic segmentation algorithm may enhance intraprostatic GTV delineation in PSMA PET images by extending the 2 main limits of conventional contouring approaches: interobserver heterogeneity and expenditure of time. Recently, convolutional neural networks (CNNs)–based algorithms achieved remarkable results handling this task. In a work by Zhao et al., the pelvic PCa tumor burden in 68Ga-PSMA PET images was detected by a CNN with 99% precision (11). Although several works already reported the excellent performance of CNNs in prostatic gland delineation on CT images (12), the use of CNNs for intraprostatic GTV contouring in PSMA PET was not examined yet. The aim of this work is to examine the capabilities of CNNs for intraprostatic GTV contouring in 68Ga- and 18F-PSMA PET.
MATERIALS AND METHODS
Patients
Data from 209 patients with primary PCa from 3 different centers (Table 1) were included. Inclusion criteria were histologically proven adenocarcinoma of the prostate and no treatment before PSMA PET. The institutional review boards approved this retrospective study, and the requirement to obtain written consent was waived.
Patient Characteristics
PET/CT Imaging
A detailed description of the radiolabeling protocol of 68Ga-PSMA-11 and 18F-PSMA-1007 from centers 1–3 can be found in previous studies (6,13⇓–15). One-hour (68Ga-PSMA-11) and 2-h (18F-PSMA-1007) after intravenous tracer injection, all patients underwent whole-body PET scanning. In center 1, protocols were acquired on 3 cross-calibrated Philips scanners: GEMINI TF TOF64, GEMINI TF16 Big Bore, and Vereos. All scanners resulted in a PET image with a voxel size of 2 × 2 × 2 mm. Center 2 used an uMI 780 PET/CT scanner (United Imaging Health Care) with a voxel size of 2.3 × 2.3 × 2.7 mm. Resampling was performed to obtain a PET image voxel size of 2 × 2 × 2 mm (trilinear interpolation in plastimatch, version 1.8.0) before training of the CNN. Expert contours of intraprostatic GTV and prostate contours were resampled with nearest neighbor interpolation (plastimatch, version 1.8.0). Center 3 acquired all studies using a Biograph mCT 128 Flow scanner (Siemens). PET images had a voxel size of 4.1 × 4.1 × 5 mm. Testing was performed with the original data and with 3 different resampling methods to obtain a PET image voxel size of 2 × 2 × 2 mm.
Histopathology and PET/CT Coregistration
For 29 patients from center 1 (cohort 1: n = 18 and cohort 2: n = 11), the 3-dimensional (3D) distribution of the intraprostatic GTV was obtained by histology information from prostatectomy specimens. The resected specimen underwent an ex vivo CT scan in customized localizer and whole-mount step sections were cut every 4 mm using a cutting device. Staining with hematoxylin and eosin was performed, and PCa tissue in histology was delineated. Histology slices were registered on ex vivo CT images, and PCa contours were transferred onto the CT images. The contours were interpolated to create a model of the 3D distribution of PCa in histology (GTV-Histo). Ex vivo CT (including GTV-Histo) was manually registered to in vivo CT. First, the prostate was delineated in both. Subsequently, ex vivo CT was oriented in the space of the in vivo CT, and the axes between the apex and the prostatic base in both CTs guided further registration. Rotation was applied for final alignment. The delineations of the prostatic glands in both CTs and intraprostatic markers (e.g., calcifications) served as reference points for anisotropic scaling of the ex vivo prostate. All coregistration steps were performed using MITK (German Cancer Research Center; version 2014.10.00).
Contouring of PSMA PET/CT
All GTVs on PET were delineated by 2 readers (GTV-Exp) from center 1 in consensus as proposed previously (10): GTVs were delineated manually in every single slice using inverted gray color scale for display, windowed with SUVmin-max: 0–5. In the first step, 2 readers with approximately 1.5 y of experience delineated the GTVs under the consideration of the respective PET/CT report. Subsequently, a reader (experience 6 y) reviewed all GTVs independently. In the case of discrepancies, each individual case was discussed and corrected to reach a consensus contour. Additionally, for the patients with histopathology reference in cohorts 1 and 2, threshold-based contouring with 30% of intraprostatic SUVmax was applied (GTV-30%) as proposed previously (16). GTV-30% volumes were created semiautomatically in Eclipse (Varian Medical Systems, USA; version 15.6). Manual contouring of the prostatic gland on CT scans was considered as the gold standard and was done using The European SocieTy for Radiotherapy and Oncology–The Advisory Committee for Radiation Oncology Practice guidelines (17). All manual delineations were created in 3D Slicer (Slicer; version 4.10.0).
Preprocessing
The data (nearly raw raster data format, nrrd) was cropped to a size of 64 × 64 × 64 voxels and normalized with 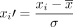 , where
, where  is the PET data of patient
is the PET data of patient 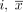 the arithmetic mean, and σ the SD within all cropped datasets. The volume of 64 × 64 × 64 voxels proved to be large enough to encompass the prostate and its surrounding tissue for all patients and small enough to enable a computation of the whole volume on the GPU.
the arithmetic mean, and σ the SD within all cropped datasets. The volume of 64 × 64 × 64 voxels proved to be large enough to encompass the prostate and its surrounding tissue for all patients and small enough to enable a computation of the whole volume on the GPU.
Because of renal excretion, it is not always possible to accurately differentiate between prostatic tissue and bladder signal in 68Ga-PSMA PET. Consequently, only delineations inside the prostatic gland contour were used for computations.
To investigate the impact of a voxel size different from the training voxel size and the usage of different interpolation algorithms, we used the PETs from center 3 in 4 different ways. First, the original data were fed to our network. In a second setting, the datasets were resampled to a resolution of 2 × 2 ×2 mm with 3 different methods (SimpleITK, version1.2.4): B-spline interpolation order 3, tri-linear interpolation and gaussian interpolation. Prostate contours and ground truth GTVs were resampled with nearest neighbor interpolation.
CNN
The current work was based on a 3D variant of the U-Net architecture (18). It consists of 3 down sampling steps with max-pooling, 3 up sampling steps with transposed convolution layers (kernel size: 2 × 2 × 2, stride: 2, padding: 1), and skip connections by concatenation. The 18 convolution blocks consist of 3 × 3 × 3 convolutions with stride and padding of 1, followed by batch normalization and rectified linear unit activation, except for the last convolution where 1 × 1 × 1 convolution without padding, batch normalization and sigmoid activation function were used. An argmax function over the final feature map formed the predicted GTV. The network weights were optimized using adaptive moment estimation (19).
Training
The 152 patients in the training cohort were further split into training (n = 142) and evaluation cohorts (n = 10). The evaluation cohort was used for optimizing the CNN’s hyper-parameters during the training process. As input the CNN received a concatenation of the patients’ PET and prostate contour. Hyper-parameter optimization was done using a grid search considering: optimizer, learning rate, number of epochs, data augmentation with x-axes flipping and scaling in x-/y-/z-direction. The best performing setting was achieved with adaptive moment estimation  a learning rate of 0.0001, and training for 1,019 epochs (an epoch means iterating over all training samples once) with a dice loss:
a learning rate of 0.0001, and training for 1,019 epochs (an epoch means iterating over all training samples once) with a dice loss:  for
for  number of labels, N image elements
number of labels, N image elements  , and without weighting the label classes
, and without weighting the label classes 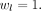 A grid search was performed without or with data augmentation by flipping the x-axis by 50% chance, by scaling the data in all directions, or by doing both. For each iteration, the original data were pseudorandomly and independently scaled in x-/y-/z-direction for ±10 voxels and then cropped as described before. Data augmentation achieved results worse than or equal to the settings without augmentation. Consequently no data augmentation was used for further analyses. In Figure 1 visualizations of the training and evaluation curves are presented.
A grid search was performed without or with data augmentation by flipping the x-axis by 50% chance, by scaling the data in all directions, or by doing both. For each iteration, the original data were pseudorandomly and independently scaled in x-/y-/z-direction for ±10 voxels and then cropped as described before. Data augmentation achieved results worse than or equal to the settings without augmentation. Consequently no data augmentation was used for further analyses. In Figure 1 visualizations of the training and evaluation curves are presented.
Visualization of the training and evaluation curves. Shown are training and evaluation results as dice loss and DSC (A) and HD and ASSD (B).
Evaluation
We assessed the agreement between GTV-Exp and GTV-CNN at voxel level using the DSC. Additionally, we considered the Hausdorff distance (HD) and the average symmetric surface distance (ASSD). The sensitivity and specificity for all GTVs based on the histology standard of reference data were calculated as performed previously (20). The prostate in each CT slice (PSMA PET/CT scans) was divided into 4 equal segments, and the analysis was performed visually using the GTVs obtained. A median of 52 segments (range: 20–64) were analyzed per patient.
Implementation
The network was implemented with pytorch 1.3.1 and torchvision 0.4.2. Gradients for backpropagation were calculated with the pytorch autograd library, which keeps track of all operations and builds a computational tree (provided code: https://gitlab.com/dejankostyszyn/prostate-gtv-segmentation).
Statistical Analysis
The statistical analysis was performed with MedPy’s package Metric Measures (version 0.4.0) and GraphPad Prism (version 8.1.0; GraphPad Software). Pairwise comparisons were performed with the Wilcoxon matched-pairs signed-rank or Friedman test. Nonpairwise testing was performed with a Mann–Whitney test or χ2 test. The tests were chosen because of nonnormal distribution (Shapiro-Wilk test) of the data. Finally, we searched for clinical factors that might impact the CNN performance by influencing the SUV distribution (PSA and Gleason score) or by neighborhood to the bladder (localization): a binary logistic regression analysis was performed to assess the impact of clinical parameter on DSC between GTV-Exp and GTV-CNN. The confidence α was set to 5%.
RESULTS
Test Results for 68Ga-PSMA-11 PET
On the internal datasets (cohort 1), the network yielded median DSC, HD, and ASSD of 0.84 (range: 0.32–0.94), 4 mm (range: 1.41–10 mm), and 0.61 mm (range: 0.24–1.46 mm), respectively (Supplemental Table 1; supplemental materials are available at http://jnm.snmjournals.org). When histology was used as a reference (Fig. 2), median sensitivity and specificity of 0.98 (range: 0.38–1) and 0.76 (range: 0.13–1) were observed. The achieved sensitivity and specificity were comparable to GTV-Exp and GTV-30% (Fig. 3). The median volumes of the GTVs were 10.7 mL (range: 0.7–101 mL) for GTV-CNN, 11.8 mL (range: 0.8–75 mL) for GTV-Exp, 8 mL (range: 2.2–41 mL) for GTV-30%, and 10.4 mL (range: 1.6–103 mL) for GTV-histo. No significant differences between absolute volume of GTV-CNN and the 3 other volumes were observed (P > 0.05). The GTV-CNN encompassed a median 26.6% of the prostatic gland.
Histology reference projected on 68Ga-PSMA-11 (upper row) and 18F-PSMA-1007 (lower row). (A and C) Hematoxylin and eosin whole-mount prostate slide with marked PCa lesion. (B and D) Axial PET image (image windowing: SUVmin-max = 0–5). Blue contour = prostate; green contour = histology reference; yellow contour = GTV-Exp; yellow contour = GTV-CNN.
Specificity and sensitivity of GTV-CNN, GTV-Exp, and GTV-30% based on comparison with histology reference. (Upper row) Cohort 1 (68Ga-PSMA-11 PET). (Lower row) Cohort 2 (18F-PSMA-1007 PET). Box plots are presented. Pairwise comparison was performed with Wilcoxon signed-rank test. n.s. = not significant. *P = 0.05–0.01.
Patients in the external test cohort (cohort 3) had statistically significant differences between Gleason scores but not between PSA values and cT stage (Table 1). Comparison between GTV-CNN and GTV-Exp was performed first on nonresampled and second on resampled PET images (Supplemental Table 1). A Friedman test revealed statistically significant (P < 0.01) differences in DSC, HD, and ASSD among the preprocessing procedures and no preprocessing. Post hoc analyses revealed no statistically significant differences between the 3 interpolation approaches (P > 0.05). As datasets with trilinear interpolation from center 2 were used in the training cohort, we conducted an additional experiment by training the CNN solely on patients from center 1 (without interpolation), to exclude a bias. Testing was performed on patients from center 3 using all 3 interpolation methods and achieved results comparable to those shown in Supplemental Table 1.
In regression analysis with pooled cohorts 1 and 3, no clinical parameter had an impact on DSC between GTV-Exp and GTV-CNN (Supplemental Table 2).
Test Results for 18F-PSMA-1007 PET
Median DSC, HD, and ASSD for cohort 2 were 0.81 (range: 0.28–0.93), 5 mm (range: 1.41–8.49 mm), and 0.51 mm (range: 0.26–1.57 mm), respectively (Supplemental Table 1). Sensitivity and specificity were 1 (range: 0.86–1) and 0.57 (range: 0.12–1). GTV-CNN had a significantly higher sensitivity than GTV-30% (P = 0.01) but not than GTV-Exp (P = 0.48). No statistically significant differences in specificity (P > 0.05) were observed between the 3 GTVs. Median volume was 3.5 mL (range: 0.3–24.4 mL) for GTV-histo, 8.5 mL (range: 1.9–38 mL) for GTV-CNN, 3 mL (range: 0.6–21.5 mL) for GTV-30%, and 7.2 mL (range: 1.2–36 mL) for GTV-Exp. GTV-CNN was statistically significantly larger (P > 0.05) than all other volumes (P < 0.05) and encompassed a median 32% of the prostate.
Computation Time
For internal test cohorts, the segmentation of the GTV of 1 patient took a median of 6 and 6.28 s, respectively, including loading and storing the data (Supplemental Table 1). This process took 23.3–27.8 s for cohort 3. A single forward pass through the CNN took less than a second (?3 μs) for all cohorts.
DISCUSSION
Implementation of automatic GTV-segmentation approaches based on CNN algorithms have already been introduced for several other tumors (21). Although several studies achieved promising results using CNNs for autosegmentation of the prostatic gland, there is limited evidence on the segmentation of the intraprostatic GTV (22). To the best of our knowledge, this is the first study analyzing CNNs for intraprostatic GTV delineation based on PET images. We chose PSMA PET images because several studies reported that PSMA PET outperformed mpMRI in tumor detection (4⇓–6). Consequently, the use of PSMA PET for initial staging (3) and intraprostatic GTV detection and contouring (23) has been established, and several studies suggested its implementation for treatment individualization in primary PCa (24⇓⇓–27). However, all previous studies used manually or semiautomatically created contours for intraprostatic GTV contouring, which may be impeded by low sensitivity or specificity and interobserver heterogeneity (10). Furthermore, manual contouring of intraprostatic GTV is time consuming. Obviously, a fast, robust and accurate workflow for intraprostatic GTV contouring is a prerequisite for a broader deployment of PSMA PET–based procedures. In this work, we proved that CNNs have the ability to delineate the intraprostatic GTV on PSMA PET with accuracy comparable to that of human experts within seconds. Thus, it is likely that PSMA PET/CT in combination with CNN-based intra- and extraprostatic (11) tumor detection and segmentation may provide a “one-stop shop” tool for tailoring individualized treatment approaches.
The CNN performance for 68Ga-PSMA PET was tested on 2 independent datasets, and high DSC values (>0.8) between GTV-Exp and GTV-CNN were observed. Bravaccini et al. reported that the PSMA expression correlates with the Gleason score (28). Because the 2 test cohorts had statistically significant differences in Gleason score in biopsy probes, our results show that the CNN performance is independent of the Gleason score and suggest that the CNN identified patterns that are independent from absolute accumulation values. Nevertheless, pattern recognition in PSMA PET images through CNNs may enable noninvasive tumor characterization (e.g., the Gleason score) in the future. In rare cases, a high HD was observed despite a high DSC. This was the case when the main parts of CNN and expert GTVs overlapped, but small regions with a high distance to the main tumor were diagnosed as malignant by the CNN, but not by the human experts. For example, in 2 patients in cohort 1 the CNN detected small (<5 mm in histology) lesions that were missed by GTV-PET. This explains the slightly higher sensitivity of the CNN in cohort 1, although the absolute GTV volumes were comparable. Because HD is sensitive to outliers, we used ASSD as an additional metric and achieved comparable results. In comparison with a histology reference GTV-CNN achieved high sensitivity and good specificity in 68Ga-PSMA PET images, which was comparable to manually delineated expert contours and threshold-based contours. Additionally, the absolute volume of GTV-CNN was similar to the histology reference volume, suggesting an adequate coverage of the intraprostatic tumor. Because GTV-CNN encompassed a median 26.6% of the prostatic gland, it is likely that focal therapy approaches guided by CNN are feasible in most of the patients. 68Ga-PSMA PET images of the external test cohort were tested with and without previous resampling. Statistically significant differences were observed with better results for the resampled datasets. Hence, when datasets from different institutions are used, a resampling of the images to the same voxel size of the training dataset should be performed. Although trilinear interpolation showed a slightly better performance, there was no statistically significant difference between the results of the 3 methods. Therefore, no specific interpolation method can be recommended. It is noteworthy that in some patients with 68Ga-PSMA PET, discrepant results between CNN and the other contours were observed. Moreover, PET signal from the adjacent bladder may mislead the CNN in contouring of PCa lesions in the prostatic base. Because no clinical parameters such as Gleason score or tumor localization had an impact on the concordance between GTV-Exp and GTV-CNN, a visual control of the CNN segmentations has to be performed for every patient.
The CNN also provided a high concordance with expert contours (DSC > 0.8) in contouring of 18F-PSMA PET images. When the differences in physical properties and in biodistribution between both tracers are taken into account, this result is surprising and should be interpreted with caution because no validated approach for contouring was applied. However, when histology as a standard of reference was considered, an excellent sensitivity was observed, which was comparable to manual contours and better than threshold-based contours. The specificity of GTV-CNN was low, which is mainly explained by a significant overestimation of the tumor volume. Thus, the CNN may also be used for GTV contouring in 18F-PSMA PET images, especially in situations in which a complete coverage of the intraprostatic GTV is demanded and a high coverage of non–tumor-bearing prostatic tissue is negligible. Surely, further studies implementing 18F-PSMA PET images and validated expert contours for training and testing are necessary to confirm this observation.
A limitation of our study is the relatively low number of patients used for testing, which is explainable by the elaborate coregistration protocol. We assume that the observed results are robust, since we used different, independent datasets for evaluation and received comparable results. Another point that supports the robustness is that we did not notice any overfitting in the training process (Fig. 1), which was further reduced by hyper-parameter optimization in combination with splitting the training data internally. Considering the high value of the 2 independent datasets used for testing the CNN, no additional approaches for validation were performed (e.g., k-fold cross-validation). Another issue is the uncertainty in correlation of PSMA PET images and histopathology slices. Thus, it could not be excluded that low coverage of PCa in histology by the PET-derived GTVs is a consequence of mismatch in coregistration or incomplete histopathologic coverage. However, as the calculation of sensitivities and specificities was not performed on a voxel-level but on a less stringent slice-by-slice level, we consider the potential resulting bias negligible. In our study, the prostatic gland on CT scans was delineated manually. Subsequent projects should integrate already existing approaches (12) for automatic prostate segmentation with our approach for automatic GTV delineation, enabling a fully automated workflow.
CONCLUSION
Our study presents a CNN for automated contouring of intraprostatic GTV in 68Ga- and 18F-PSMA PET. Likewise, CNN-based GTV delineation is a promising and fast alternative to visual and threshold-based PET image interpretation. The link to the code and trained model of the CNN may be used for focal therapy or targeted biopsy concepts in primary PCa by providing a GTV proposal before visual image interpretation. We strongly emphasize that our tool is not clinically validated and not certified, thus a visual control of the CNN contours by experienced experts is obligatory. Furthermore, the CNN may be used as an alternative approach for GTV segmentation in ongoing radiomics or deep learning research in the field where certification is not mandatory.
DISCLOSURE
This study was funded from the ERA PerMed call 2018 (BMBF). No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: How is the performance of a trained CNN for automatic segmentation of intraprostatic tumor volume in PSMA PET images of primary prostate cancer patients?
PERTINENT FINDINGS: In this multicenter study including 209 patients, the CNN provided results comparable to those by human experts and threshold-based delineations and coregistered whole-mount sections as the standard of reference were considered.
IMPLICATIONS FOR PATIENT CARE: The CNN provided a fast and robust auto-segmentation of the intraprostatic tumor and may enhance individualized therapeutic approaches for primary prostate cancer patients such as focal therapy or targeted biopsy.
Footnotes
- COPYRIGHT © 2021 by the Society of Nuclear Medicine and Molecular Imaging.
REFERENCES
- Received for publication July 31, 2020.
- Accepted for publication October 7, 2020.










