Abstract
We propose a new deep learning–based approach to provide more accurate whole-body PET/MRI attenuation correction than is possible with the Dixon-based 4-segment method. We use activity and attenuation maps estimated using the maximum-likelihood reconstruction of activity and attenuation (MLAA) algorithm as inputs to a convolutional neural network (CNN) to learn a CT-derived attenuation map. Methods: The whole-body 18F-FDG PET/CT scan data of 100 cancer patients (38 men and 62 women; age, 57.3 ± 14.1 y) were retrospectively used for training and testing the CNN. A modified U-net was trained to predict a CT-derived μ-map (μ-CT) from the MLAA-generated activity distribution (λ-MLAA) and μ-map (μ-MLAA). We used 1.3 million patches derived from 60 patients’ data for training the CNN, data of 20 others were used as a validation set to prevent overfitting, and the data of the other 20 were used as a test set for the CNN performance analysis. The attenuation maps generated using the proposed method (μ-CNN), μ-MLAA, and 4-segment method (μ-segment) were compared with the μ-CT, a ground truth. We also compared the voxelwise correlation between the activity images reconstructed using ordered-subset expectation maximization with the μ-maps, and the SUVs of primary and metastatic bone lesions obtained by drawing regions of interest on the activity images. Results: The CNN generates less noisy attenuation maps and achieves better bone identification than MLAA. The average Dice similarity coefficient for bone regions between μ-CNN and μ-CT was 0.77, which was significantly higher than that between μ-MLAA and μ-CT (0.36). Also, the CNN result showed the best pixel-by-pixel correlation with the CT-based results and remarkably reduced differences in activity maps in comparison to CT-based attenuation correction. Conclusion: The proposed deep neural network produced a more reliable attenuation map for 511-keV photons than the 4-segment method currently used in whole-body PET/MRI studies.
Although PET/MRI is an emerging hybrid imaging modality (1–4), the PET attenuation maps (μ-maps) used in whole-body PET/MRI studies have limited accuracy (5,6). In brain PET/MRI studies, attenuation correction has advanced greatly through the use of pseudo CT images generated by segmenting ultrashort echo time or zero echo time MR images and by registering an atlas generated from transmission or CT scans to individual data (7–9). However, these techniques used in brain PET/MRI have not been successfully applied in whole-body PET/MRI studies. The Dixon sequence–based 4-segment approach used as a standard method in whole-body PET/MRI systems (10) underestimates PET activity in bone structures because of the lack of bone information in attenuation maps (11,12). Although a model-based approach (13) has been suggested to add bone structures to 4-segment maps (14), inaccurate registration between PET images and bone models may cause errors in PET activity quantification.
Although algorithms for simultaneous reconstruction of activity and attenuation have been greatly improved by the incorporation of time-of-flight (TOF) information (15,16), their accuracy is still far from the clinically relevant level. The maximum-likelihood reconstruction of activity and attenuation (MLAA) algorithm, a simultaneous reconstruction algorithm, has the advantage of providing attenuation maps (17,18). MLAA-generated attenuation maps allow the use of image-domain priors to improve the algorithm accuracy and convergence (19,20). However, mainly because of the insufficient timing resolution of clinical PET systems, the MLAA suffers from slow convergence, high noise levels in attenuation maps, and crosstalk between activity and attenuation distribution (21).
Deep learning–based approaches have been suggested to improve the accuracy of regional PET attenuation correction. In our recent work (22), to mitigate the limitations of MLAA in brain PET, deep convolutional neural networks (CNNs) were trained to learn a true CT-derived attenuation map with the MLAA activity and attenuation maps as their inputs. The CNNs generated less noisy and more uniform attenuation maps than original MLAA, resulting in only 5% errors in activity and binding ratio quantification in the most challenging brain PET cases for simultaneous image reconstruction (dopamine transporter imaging). Another notable application of deep learning for this purpose is CNN-based PET attenuation map generation from the zero echo time and Dixon MRIs (23). In pelvis PET/MRI studies, this multiparametric MRI–based approach reduced the PET quantification error in bone lesions by a factor of 4 in comparison to the conventional Dixon sequence–based 4-segment approach. However, this approach requires additional zero echo time MRI acquisition with a relatively long scan time.
In this study, we investigated the feasibility of the deep learning–based approach to whole-body PET/MRI attenuation correction without use of zero echo time or ultrashort echo time data. Similarly to our previous work on the brain PET studies (22), we use MLAA-based activity and attenuation maps as inputs to CNN to learn a CT-derived attenuation map. However, we conducted 3-dimensional (3D) patch-based learning instead of 2-dimensional slice-by-slice mapping because whole-body structures are more complex than head region and 3D learning allows better continuity of image intensity in the axial direction. The CNN was trained and tested using oncologic whole-body 18F-FDG PET/CT scan data. Then, the similarity of the CNN-generated attenuation map and the CT-derived map was evaluated. Also, the attenuation-corrected PET images produced using the conventional 4-segment method and new CNN outcomes were compared with ground truth (CT).
MATERIALS AND METHODS
Subjects and Image Acquisition
The whole-body 18F-FDG PET/CT scan data of 100 cancer patients (38 men and 62 women; age, 57.3 ± 14.1 y) acquired using a Biograph mCT 40 scanner (effective timing resolution, 580 ps; Siemens Healthcare) from March 2017 to May 2017 were retrospectively analyzed. The retrospective use of the scan data and waiver of consent were approved by the Institutional Review Board of our institute. For all patients, PET/CT imaging was performed 60 min after intravenous injection of 18F-FDG (5.18 MBq/kg). The patients’ upper bodies from head to upper thigh were covered by a 6- to 8-bed-position emission scan (scan time, 1 min/bed position).
Sinograms of prompt PET counts and correction factors were generated using the e7 toolkit. The CT images were reconstructed in a 512 × 512 × 100 matrix with the voxel size of 1.52 × 1.52 × 2.03 mm and converted into the μ-map for 511-keV photons (200 × 200 × 109; 4.07 × 4.07 × 2.03 mm). We reconstructed PET datasets using the MLAA with the TOF information (6 iterations and 21 subsets, 5-mm gaussian postprocessing filter). The matrix size of MLAA-reconstructed images was 200 × 200 × 109 (4.07 × 4.07 × 2.03 mm voxel size) for each bed position.
Simultaneous Activity and Attenuation Reconstruction Algorithm
In each iteration n, a standard maximum-likelihood expectation maximization algorithm updates activity distribution λ for a current attenuation coefficient μ as follows:
 where
where  is the length of intersection between LOR i and voxel j, and
is the length of intersection between LOR i and voxel j, and  is the TOF system matrix element between LOR i, TOF bin t, and voxel j. Also, y is the measured emission projection and s is the additive correction factor that contains scatter and random events.
is the TOF system matrix element between LOR i, TOF bin t, and voxel j. Also, y is the measured emission projection and s is the additive correction factor that contains scatter and random events.
After updating activity distribution, the attenuation coefficient is updated by a maximum-likelihood-for-transmission-tomography algorithm as follows:

To resolve the nonunique global scaling problem in MLAA, the boundary constraint was applied during the attenuation image estimation in MLAA, following the original TOF MLAA paper (17).
Data Set
The data of 60 patients were used for training the CNNs, that from 20 others as a validation set to prevent overfitting, and the other 20 as a test set (Table 1) for the CNN performance analysis. For the CNN training and testing, activity and attenuation maps derived from MLAA were used as input to the CNN, and an attenuation map converted from CT (μ-CT) as label data (Fig. 1). We generated the attenuation map based on a 4-segment approach (μ-segment) from μ-CT by classifying each patient’s body parts into 4 segments using 3 different thresholds applied to μ-CT. The thresholds between air, lung, fat, and water were 0.015, 0.04, and 0.09. The mean μ-value for each segment was then calculated from 60 μ-CT images and assigned to the body segment of each patient.
Demographic Data of Patients Included in Test Set
Data flow in generation of attenuation (μ) map using proposed deep neural network for whole-body PET/MRI attenuation correction and architecture of deep neural network. Deep neural network was trained to learn ground truth CT-derived μ-map from MLAA-derived activity and attenuation maps. In network architecture, each box represents feature map resulting from operation corresponding to its color. Number of features in each layer is indicated at bottom of box. Dimensions of each feature map are shown below box.
All inputs and labels for the CNN were in the form of 32 × 32 × 32 matrix patches. The intensity of each activity patch was normalized to have the range between 0 and 1. To avoid including meaningless blank patches during the CNN training, the volume patches were used for training only if their centers were included in the body. With this inclusion criterion, approximately 1.3 million patches were used for CNN training.
Network Architecture
The network was designed to predict μ-CT from activity and attenuation maps derived from MLAA. The network consists of a contracting path to capture the context and a symmetric expanding path that enables precise localization (24). As shown in Figure 1, the left half of the proposed network (contracting path) includes repeating two 3D convolution layers with rectified linear units and batch normalization, and 2 × 2 × 2 max-pooling layers for down sampling. Similarly, in the right half of the network (expanding path), two 3D deconvolution layers with rectified linear units and batch normalization are repeated. In every layer, the number of feature maps is doubled in the contracting path and reduced by half in the expanding path. In the first layer, a convolution with 3 × 3 × 3 × 2 kernels is applied to merge 2 input datasets. Then, each convolution and deconvolution layer except for the last one is composed of 3 × 3 × 3 kernels. In the last layer, which provides an output, 1 × 1 × 1 convolution is used for scaling purposes. Symmetric skip connections (copy and concatenation) between the convolution and deconvolution layers are used to achieve fast convergence and attain high frequency of local features (25). We implemented the networks using the TensorFlow library (26).
Network Training and Loss Function
The L1 norm between output (μ-CNN) and ground truth (μ-CT) was chosen as a cost function to train the network to generate a CT-like attenuation map. The cost function was minimized using adaptive moment estimation with an initial learning rate of 0.001, which was reduced by half every 2 epochs (27). In every epoch, we updated the network 9,955 times using a mini-batch of 128 samples. The total number of epochs was 10. The training time was approximately 30 h/epoch when a Ryzen 1700X central processing unit with a GTX 1080 graphics processing unit was used. Supplemental Figure 1 shows that the L1 norm between μ-CNN and μ-CT decreases as the epoch increases, and 10 epochs are sufficient to reach convergence (supplemental materials are available at http://jnm.snmjournals.org).
Image Analysis
The relative performance of CNN-based attenuation correction of whole-body 18F-FDG PET was analyzed by comparing the attenuation and activity maps generated from 20 test sets. The attenuation maps generated using the proposed method, MLAA, and 4-segment method (μ-CNN, μ-MLAA, and μ-segment) were compared with μ-CT, a ground truth. A voxelwise scatterplot was plotted for comparison, and the correlation coefficients were calculated. The similarity of segmented regions from μ-CNN and μ-MLAA attenuation maps was compared using the Dice similarity coefficient (28,29). For the segmentation of μ-CNN and μ-MLAA, the sample thresholds used for generating μ-segment were applied (0.015, 0.04, and 0.09 between air, lung, fat, and water).
Attenuation-corrected activity maps were also obtained using the ordered-subset expectation maximization (3 iterations, 21 subsets, 5-mm gaussian postprocessing filter) algorithm with different attenuation maps. We made a voxelwise comparison between the activity maps generated using the proposed method, MLAA, and 4-segment method (λ-CNN, λ-MLAA, and λ-segment) and the activity map obtained using μ-CT (λ-CT). We also compared the SUVmean of primary and metastatic bone lesions (29 lesions from 6 patients) and soft-tissue lesions (11 lesions from 5 patients) obtained by manual drawing of regions of interest on the activity images of the patients who were included in the test set and showed abnormal uptake (Table 1).
The peak signal-to-noise ratio and normalized root-mean-square error values were calculated as additional image quality metrics to quantify the similarity of attenuation and activity maps (30).
To understand better what the CNN is really doing, some sanity checks were performed by exploring the CNN output for μ-MLAA input clipped to soft-tissue attenuation values (no higher bone attenuation), λ-MLAA input with high–low flipped activity, and μ-MLAA input globally scaled by 0.95.
RESULTS
Attenuation Maps
The CNN generates less noisy attenuation maps than μ-MLAA and achieves better bone identification than μ-segment and μ-MLAA. Figure 2 shows sagittal and coronal slices of λ-MLAA, μ-MLAA, μ-segment, μ-CNN, and μ-CT in a representative case. Although μ-MLAA shows higher μ-values in bone regions than soft-tissue and lung tissue, the margin between different tissues is not as clear as in the other attenuation maps. Moreover, some soft-tissue regions (e.g., back and shoulder) show improperly high μ-values, and lower spinal bone regions are not well identified (Fig. 2B). In contrast, the large bone structures are more accurately delineated in the μ-CNN, although the small bone structures are not as fine as those obtained with μ-CT (Fig. 2D). Supplemental Figure 2 shows the attenuation maps generated from another representative case that also proves the superiority of the CNN-based approach to the others in terms of the similarity to μ-CT. However, some small air regions in the abdomen are missing in the μ-CNN (Supplemental Fig. 2D), indicating the limited performance of CNN to overcome the high noise-level and low resolution of λ-MLAA and μ-MLAA.
Attenuation maps of representative case. (A) λ-MLAA: original MLAA activity. (B) μ-MLAA: original MLAA attenuation map. (C) μ-segment: 4-segment map corresponding to Dixon MRI-based attenuation map. (D) μ-CNN: deep CNN output. (E) μ-CT: ground truth.
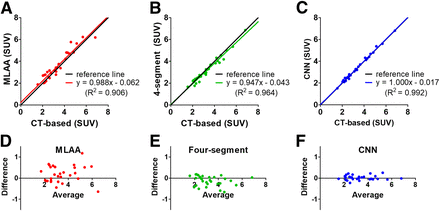
The quantitative analysis on the similarity of attenuation maps confirmed the qualitative comparison results. The μ-CNN achieved the best voxelwise correlation with μ-CT as shown in Figures 3A–3C (the joint histogram of μ-values were plotted in log scale) and Table 2. Also, the μ-CNN yielded the highest peak signal-to-noise ratio and lowest normalized root-mean-square error relative to the μ-CT (Table 3). The average Dice similarity coefficients between μ-CNN and μ-CT were significantly higher than that between μ-MLAA and μ-CT in all regions (Fig. 4). In the regions of interest drawn on the bone lesions, the μ-CNN yielded the smallest error relative to the μ-CT (MLAA, −4.10% ± 10.62%; 4-segment, −4.20% ± 8.23%; CNN, 0.43% ± 6.80%). In the soft-tissue lesions, both 4-segment method and CNN yielded considerably smaller error than MLAA (MLAA, −4.57% ± 6.59%; 4-segment, 0.57% ± 1.74%; CNN, 0.91% ± 3.55%).
(A–C) Correlation between μ-CT and μ-MLAA (A), μ-segment (B), and μ-CNN (C). (D–F) Correlation between λ-CT and λ-MLAA (D), λ-segment (E), and λ-CNN (F). Red lines are identity lines.
Summary of Voxelwise Correlation of Attenuation (μ) and Activity (λ) Relative to Ground Truth (μ-CT and λ-CT) for 20 Subjects Used in Test Set
Peak Signal-to-Noise Ratio and Normalized Root-Mean-Square Error Relative to Ground Truth (μ-CT and λ-CT) for 20 Subjects Used in Test Set
Statistical analysis on Dice similarity coefficients between μ-CNN and μ-CT and those between μ-MLAA and μ-CT.
Similar to our previous results (22), the activity information enhances the CNN performance. Supplemental Figure 3 shows the results of CNNs trained with and without λ-MLAA as input. Using both the λ-MLAA and μ-MLAA as input, we could generate μ-CNN with better image contrast and anatomic detail.
Through the sanity checks, we could verify that the CNN works properly as we intended. As shown in Figure 5, the μ-MLAA input clipped to soft-tissue attenuation values yielded μ-CNN with missing bone regions. Intensity-flipped λ-MLAA input produced irreverently homogeneous μ-CNN, and μ-MLAA input globally scaled by 0.95 led to the underestimation of μ-CNN intensity.
Attenuation maps generated in sanity checks to understand how CNN works. (A–C, top) CNN outcomes for μ-MLAA input clipped to soft-tissue attenuation values (no higher bone attenuation) (A), λ-MLAA input with high–low flipped activity (B), μ-MLAA input globally scaled by 0.95 (C). (A–C, bottom) Differences from original CNN output.
Activity Maps
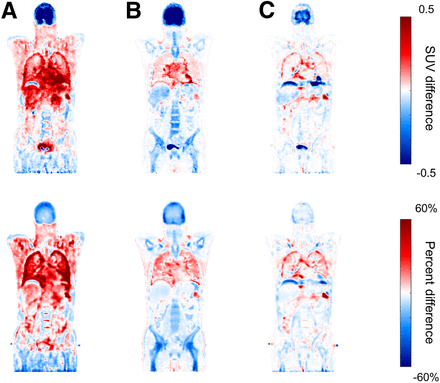
The accuracy of PET activity quantification was improved by the attenuation correction using μ-CNN. As shown in Figures 3D–3F and Table 2, the λ-CNN more strongly correlates with λ-CT than do the λ-MLAA and λ-segment. In addition, the λ-CNN showed the highest peak signal-to-noise ratio and lowest normalized root-mean-square error relative to λ-CT (Table 3). Figure 6 shows the differences in activity maps (SUV) relative to CT-based in a representative case. Although λ-MLAA (A) shows lower differences in bone regions than λ-segment (B), the differences in soft tissues were higher, particularly in the lungs, heart, liver dome, and bladder. The differences between λ-CNN and λ-CT (C) were smaller and more uniform than the others. The overestimation of activity in the lungs and underestimation in the liver seen in λ-MLAA were considerably reduced in λ-CNN. Supplemental Figure 4 shows attenuation-corrected PET images of a patient with lung lesions using different attenuation maps.
Differences of SUV between λ-CT and λ-MLAA (A), λ-segment (B), and λ-CNN (C). SUV difference = λ-method − λ-CT; percent difference = (λ-method − λ-CT)/λ-CT×100%; method = MLAA, segment, or CNN.
The same trend was observed in the regional SUVmean quantification in bone lesions (Fig. 7) and soft lesions. The relative differences in bone lesions and soft-tissue lesions are plotted in Supplemental Figure 5. Although the 4-segment map-based correction (y = 0.947x − 0.043, R2 = 0.964) improved the SUV correlation with CT-based attenuation correction in comparison to the original MLAA (y = 1.020x − 0.062, R2 = 0.848), the CNN (y = 1.000x − 0.017, R2 = 0.992) outperformed the 4-segment map-based correction. However, in 6 vertebrae regions of 29 bone lesions, MLAA (2.71% ± 4.61%) and CNN (−2.22% ± 1.77%) were more accurate than 4-segment map-based correction (−9.40% ± 5.16%). In soft lesions, CNN (1.31% ± 3.35%) showed smaller error than MLAA (8.78% ± 7.41%) and 4-segment map-based correction (−2.90% ± 1.22%). Also, the CNN yielded the lowest voxelwise average error relative to the CT-based attenuation correction (λ-MLAA, 12.82% ± 2.45%; λ-segment, 5.61% ± 0.68%; λ-CNN, 2.05% ± 1.51%).
Comparison of SUVmean measured in bone lesions (29 lesions in 10 patients in test set). Scattered plots between λ-CT and λ-MLAA (A), λ-segment (B), and λ-CNN (C) and corresponding Bland–Altman plots (D–F).
Computation Time
The MLAA took approximately 2 times longer than ordered-subset expectation maximization, but the CNN inference did not require long computation time (<30% of ordered-subset expectation maximization reconstruction for whole-body PET).
DISCUSSION
Attenuation correction is an essential procedure in the generation of PET images with quantitatively accurate regional activity information. In PET/CT systems, the Hounsfield unit in CT images is converted into the linear attenuation coefficient for 511-keV annihilation photons based on their bilinear relationship to generate patient-specific PET attenuation maps (μ-CT). However, attenuation map generation in PET/MRI is not so straightforward because the MR signal is not directly related to photon attenuation (6). The Dixon MRI based on the chemical shift difference between water and fat provides noiseless 4-segment maps for PET attenuation correction (10). However, these 4-segment maps cannot accurately account for photon attenuation by bone tissue (11,12,31). In addition, the inter- and intrapatient variability of μ-values is ignored in this method (11,32). Moreover, the bone model–based approach cannot handle heterogeneous bone attenuation coefficients even in cases with accurate bone registration. Although ultrashort echo time MRI sequence-based conversion of R2* to the CT Hounsfield unit was proposed, its application is limited to neurologic studies (29,33). Despite recent advances in the accuracy of TOF measurement in PET, limitations of simultaneous activity and attenuation reconstruction still exist in real clinical cases. The deep CNN trained in this study to learn μ-CT from MLAA activity and attenuation maps handled well the information provided by this physically relevant but imperfect attenuation correction method.
The convolution kernels of CNN, which determine how the MLAA images are merged, evolve during the network training based on the given training set. Therefore, the CNN approach does not need additional image segmentation, tissue probability prior, control parameters, and so forth (34–36), which are required in other approaches that have been previously proposed to combine MLAA and Dixon attenuation correction methods (37,38). For example, the MRI-guided MLAA algorithm (37,39), which imposes MR spatial and CT statistical constraints on the MLAA estimation of attenuation maps using a constrained gaussian mixture model and a Markov random field smoothness prior, needs to use a coregistered bone probability map. Another joint estimation algorithm that uses Dixon MRI as a prior requires uncertain MR-region segmentation and emphatically determined prior weights for this segment (38).
The difference between PET activities corrected for attenuation using proposed CNN-based method and CT was relatively high in lung boundary and liver dome (Fig. 6C). Incorrect activity measurement in lung and upper liver due to the respiratory motion–induced mismatch between PET and CT data is a well-known artifact in CT-based PET attenuation correction. In the MLAA, the activity and attenuation are simultaneously estimated only from the emission PET data without use of any transmission data. Therefore, the MLAA activity and attenuation maps would be free from or less vulnerable to the position mismatch artifact. We used CT as the ground truth for training the CNN. However, major data used for network training came from other regions rather than lung and upper liver because we performed patch-based learning. In fact, the λ-MLAA, μ-MLAA, and μ-CNN show well-matched boundaries of liver dome (Figs. 2A, 2B, and 2C) although the liver dome in μ-CT is elevated (Fig. 2E). Accordingly, the CNN-based approach that derived an attenuation map from the MLAA outputs would also be less influenced by the respiratory motion than CT-based attenuation correction, resulting in the activity difference shown in Figure 6C.
A limitation of this study is that we trained and validated the CNN only for whole-body 18F-FDG PET scans. It is uncertain yet whether the trained deep network for simultaneously reconstructed activity and attenuation maps from 18F-FDG will work for other types of PET radiotracers. Further investigation is required to answer this clinically important question. Even if different network parameters are required for each type of radiotracer, the network parameters derived from 18F-FDG PET in this study could serve as initial values assigned before fine parameter tuning for each radiotracer. This technique, called transfer learning, allows faster convergence in training with a limited training set (40). The transfer learning will also be useful for training the CNN when it is applied to the PET systems with different timing resolutions. Another direction to advance in future investigations would be the combination of MLAA outputs and Dixon MR images as the input to the CNN. The detailed anatomic information and tissue characterization provided by the Dixon MR images would be useful for improving the CNN performance. It should also be noted that the performance of MLAA is sensitive to accurate scatter estimation and TOF timing offset calibration (41). Thus also, the CNN MLAA performance would be influenced by the residual uncertainties in scatter estimation and TOF timing offset calibration.
CONCLUSION
We have developed a deep neural network that successfully processes the information obtained from simultaneous activity and attenuation reconstruction to produce a more reliable attenuation map for 511-keV photons in comparison to the conventional 4-segment method. We also verified its feasibility using a whole-body 18F-FDG PET dataset with TOF information. Accordingly, the proposed method has potential to replace the current 4-segment–based attenuation correction in whole-body FDG PET/MRI in which bones are poorly identified in whole-body PET/MRI studies and the local MRI signal loss produced by metallic implants results in the considerable error in image segmentation. Also, we expect that the accuracy of this new method will improve as the TOF and machine learning technologies advance further.
DISCLOSURE
This work was supported by grants from the National Research Foundation of Korea (NRF) funded by the Korean Ministry of Science and ICT (NRF-2014M3C7034000, NRF-2016R1A2B3014645, and NRF-2017M3C7A1044367). The funding source had no involvement in the study design, collection, analysis, or interpretation. No other potential conflict of interest relevant to this article was reported.
Footnotes
Published online Jan. 25, 2019.
- © 2019 by the Society of Nuclear Medicine and Molecular Imaging.
REFERENCES
- Received for publication August 28, 2018.
- Accepted for publication December 20, 2018.