Abstract
The mouse double-minute 2 (MDM2) oncogene, amplified or overexpressed in many human cancers, has been suggested to be a novel target for cancer therapy. Visualization of MDM2 expression using radionuclide targeting can provide important diagnostic information in malignant tumors. The overall aim of this study was to evaluate whether liposome-coated 99mTc-radiolabeled antisense oligonucleotides (ASONs) targeting MDM2 messenger RNA (mRNA) could be used for imaging of MDM2 expression in vivo. Methods: ASON and mismatch oligonucleotide (ASONM) targeted to MDM2 mRNA were synthesized and radiolabeled with 99mTc using the bifunctional chelator hydrazinonicotinamide (HYNIC). Then the radiolabeled probe was characterized in vitro. Reverse-transcriptase polymerase chain reaction and Western blotting were performed to assay the MDM2 mRNA and protein level after MCF-7 (human breast adenocarcinoma cell line) cells were incubated with liposome-coated 99mTc-HYNIC-ASON/ASONM at various concentrations for 24 h. Following established MCF-7–bearing nude mice models, the biodistribution of liposome-coated 99mTc-HYNIC-ASON/ASONM was investigated, and in vivo tumor scintigraphic images were acquired for these animal models. All data were analyzed by statistical software. Results: The labeling efficiencies of 99mTc-HYNIC-ASON and 99mTc-HYNIC-ASONM were 57.2% ± 2.98% and 56.3% ± 3.01%, respectively; the specific activities were 1,450 ± 60.2 and 1,370 ± 55.4 kBq/μg, respectively; and the radiochemical purity for both was above 95%. The radiolabeled ASON still had the ability to hybridize to the sense oligonucleotide. In comparison with the mismatch probe, the antisense probe had an obvious effect on the levels of MDM2 mRNA and protein. The levels of mRNA and protein were significantly different for different concentration antisense probe groups (P < 0.01). The excretion of the antisense and mismatch probe was mainly through the liver and kidneys. The tumor radioactivity uptake of the antisense probe was significantly higher than that of the mismatch probe (P < 0.01). At 1–10 h after injection of the antisense probe, the tumor could be clearly visualized, whereas the tumors were not imaged at any time after injection of the mismatch probe. Conclusion: The accumulation of liposome-coated 99mTc-labeled ASONs in breast cancer tissue is specific. The antisense imaging with liposome-coated 99mTc-HYNIC-ASON may be a promising method for visualization of MDM2 expression in human breast cancer.
Mouse double-minute 2 (MDM2), the cellular ubiquitin E3 ligase of the tumor suppressor p53, is considered to be an oncoprotein because of its activity in promoting p53 ubiquitination and proteasomal degradation. The activation of p53 target genes induces apoptosis, cell cycle arrest, and senescence, which are important to tumor suppression (1). In normal cells, p53 and MDM2 mutually form a negative-feedback return route and are precisely balanced (2). DNA damage in cells with higher levels of p53 activates MDM2 gene transcription and increases its protein expression levels. MDM2 protein binds to the p53 transcriptional activator protein-specific functional areas, inhibits the activation of transcription functions, and facilitates the degradation of p53 protein by the ubiquitin-dependent pathway, resulting in irreversible deactivation of p53. The negative-feedback return route to the growth inhibitory activity of p53 can be limited to a reasonable level. This is a normal physiologic function of organisms to maintain their means of balance control. However, in some cells, gene amplification or other causes increase levels of MDM2, resulting in the nonnormal inhibition of p53 and loss of control cell growth, thereby changing the characteristics of tumor cells (3).
Recently, p53-independent tumorigenic mechanisms for MDM2 have been identified (4). MDM2 also binds to other proteins with a wide range of functions. For example, MDM2 promotes cell cycle progression by binding to and modulating the activities of p21Waf1/CIP1 (5,6) and E2F1 proteins (7).
Studies have demonstrated that MDM2 overexpression is associated with advanced cancer disease states, such as invasive (8–10) and high-grade or late-stage tumors (11), recurrence (12), and metastasis (13,14). In addition, because overexpression of MDM2 in human cancers is associated with a poor prognosis (15), MDM2 is considered to be a target for human cancer therapy (16–20).
Antisense imaging is one of the most promising tools of molecular imaging, whose strategies exploit the exquisite specificity of nucleic acid base-pair (bp) binding. Compared with the traditional imaging techniques for diagnosing breast cancer, such as MRI, computer tomography, ultrasound imaging, and mammography, antisense imaging is attractive in that it detects the expression of specific genes rather than the anatomy variance. Noninvasive, real-time imaging of oncogene expression in vivo would reveal molecular changes in diseased tissues at relatively early stages. Moreover, high specificity due to sequence complementarity may be achieved on binding of the radiolabeled oligonucleotides to the target. However, in antisense imaging there are challenges, such as in vivo stability, transport to the target, entry into the cell, and hybridization with target-specific sequences. The development of antisense imaging agents, compared with other approaches of molecular imaging, is in its infancy. However, antisense imaging for early, specific, and noninvasive detection of oncogene expression is unique and warrants greater attention.
Considering the role of MDM2 in the development and treatment of cancer, in the present study we tried to use antisense oligonucleotides (ASONs) to noninvasively measure MDM2 messenger RNA (mRNA) expression directly. One 20-mer antisense uniformly phosphorothioate-modified oligonucleotide targeting MDM2 mRNA was radiolabeled with 99mTc. We evaluated the characteristics of this antisense probe in vitro and observed whether it could be used for imaging of MDM2 expression in human breast cancer in vivo.
MATERIALS AND METHODS
Three single-strand 20-mer DNA oligonucleotides were purchased from Shanghai Sangon Biologic Engineering Technology and Services Co., Ltd. The ASON sequence was 5′-GATCACTCCCACCTTCAAGG-3′, the sense oligonucleotide sequence was 5′-CCTTGAAGGTGGGAGTGATC-3′, and the ASONM sequence was 5′-GATGACTCACACCATCATGG-3′ (the italicized nucleosides were the sites of the mismatch). For these 3 oligonucleotides, all internucleotide linkages were phosphorothioate. Every DNA had a primary amine on the 5′ end, attached through a 6-carbon alkyl linker.
Hydrazinonicotinamide (HYNIC) was synthesized and purified at the Department of Chemistry at Beijing Normal University. 99mTcO4− was obtained from a 99Mo–99mTc radionuclide generator made by the China Institute of Atomic Energy. N,N-dimethylformamide, tricine, and Sncl2·2H2O were supplied by Sigma, and Trizol was provided by Gibco Co. The antihuman MDM2 (SMP-14) and p53 (DO-1) monoclonal antibody were purchased from Santa Cruz Biotechnology. Lipofectamine 2000 was purchased from Invitrogen Co. Sep-Pak C18 reverse and Sephadex G25 columns were obtained from Waters Co.
Oligonucleotide Conjugation and Radiolabeling
The solution of DNA (5 μg/μL) in 25 mM bicarbonate buffer (pH 8.5) was added dropwise to HYNIC in dry N,N-dimethylformamide (10 mg/mL) until a molar ratio of 20:1 (HYNIC:DNA) was reached. After the solution was incubated at room temperature for 60 min, the coupling mixture was purified by Sep-Pak C18. Briefly, the Sep-Pak C18 column was washed with 10 mL of high-performance liquid chromatography–grade acetonitrile and rinsed with a 20-mL aliquot of sterile filtered water. The coupling reaction mixture was diluted to 1 mL with water, and the entire sample was applied to the column. The column was washed with the following 3 solutions: 10 mL of 25 mM ammonium bicarbonate (pH 8.5), 10 mL of 25 mM ammonium bicarbonate/5% acetonitrile, and 2 × 10 mL of water/5% acetonitrile. The DNA was then eluted with 4 × 1 mL of water/30% acetonitrile. One-milliliter fractions were collected and quantified at 260 nm by ultraviolet spectrophotometry. The fractions containing DNA were pooled and dispensed at 10 μg/vial, dried with a centrifugal evaporator, and stored at −20°C for future use.
Tricine was used in this study as a coligand for HYNIC. Solid HYNIC-DNA (10 μg) was dissolved in 20 μL of 25 mM bicarbonate buffer, pH 8.5. HYNIC-DNA solution was added to 100 μL of tricine solution (7 mg/mL in water), 74–185 MBq (10–20 μL) of 99mTc-pertechnetate solution, and a fresh 5.5 μL of SnCl2·2H2O solution (1 mg/mL in 0.1 M HCl). After the solution was incubated for 60 min at room temperature, the 99mTc-HYNIC-DNA was purified by Sephadex G25 column. The fractions were collected to detect the radioactivity and absorbance at 260 nm. The labeling efficiency and specific activity were then assayed.
HYNIC-DNA (10 μg) was radiolabeled, added to the reversed-phase Sep-Pak C18 column, and then eluted with acetonitrile solution (30% in 0.1 M ammonium acetate). Radioactivities and absorbance at 260 nm were detected to identify and quantify the peak fractions. Radioactivity of the Sep-Pak C18 column was also calculated for the radiochemical purity assessment.
Molecular Hybridization Activity
After the unlabeled sense oligonucleotide was added to 99mTc-HYNIC-ASON according to a molar ratio of 1:1 at 37°C, aliquots were removed for reversed-phase Sep-Pak C18 column chromatography. Compared with the result of proincubation, the radioactivity peak diversion was surveyed.
Cell Culture
The MCF-7 cell line was a gift from the Tumor Hospital of Harbin Medical University. MCF-7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 100 mg of penicillin–streptomycin per milliliter. For the following studies, MCF-7 cells were detached by trypsin and counted and then plated in 6-well flat-bottomed culture plates in DMEM with 10% fetal bovine serum (2 mL/well, 2.5 × 106 cells per well) and incubated at 37°C, 50 mL/L CO2 for 24 h, until the cell density reached 90%.
Cell Transfections
After the 10 μg of purified 99mTc-HYNIC-DNA were diluted in 500 μL of DMEM and 25 μL of Lipofectamine 2000 were diluted in 475 μL of DMEM, the 2 mixtures were combined and incubated for 20 min at room temperature. Thus, the DNA concentration is 0.01 μg/μL.
After MCF-7 cells were incubated with the control group of 10% fetal bovine serum, liposome-coated 99mTc-HYNIC-ASON (100–500 nmol/L), and 99mTc-HYNIC-ASONM (500 nmol/L) for 6 h at 37°C, the medium in 6-well plates was removed and changed to DMEM with 10% fetal bovine serum. After 24 h of incubation at 37°C, MCF-7 cells were collected, frozen (−20°C), and melted 3 times for future use.
Reverse-Transcriptase Polymerase Chain Reaction (RT-PCR)
The mRNA levels in cells incubated with liposome-coated 99mTc-HYNIC-ASON and 99mTc-HYNIC-ASONM were analyzed by RT-PCR. Total RNA was extracted using the Trizol reagent, and cDNA was generated using the RT-PCR kit from Invitrogen. PCR coamplifications of MDM2, p53, and internal reference β-actin were performed. The MDM2 primers were 5′-AATCATCGGACTCAGGTACA-3′ for sense and 5′-GTCAGCTAAGGAAATTTCAGG-3′ for antisense, and the product was 564 bp. The p53 primers were 5′-CCTTCCCAGAAAACCTACCA-3′ for sense and 5′-TCATAGGGCACCACCACACT-3′ for antisense, and the product was 371 bp. The β-actin primers were 5′-ACACTGTGCCCATCTAACGAGG-3′ for sense and 5′-AGGGGCCGGAACTCGTCATACT-3′ for antisense, and the product was 621 bp. PCR products were separated and virtualized on 0.8% agarose gel electrophoresis. Every band of PCR product was scanned using a gel imaging system. Considering β-actin as the inside reference, quantitation of mRNA were demonstrated with the ratios of specific band and inside analyze reference.
Western Blotting
The protein levels of MDM2 and p53 were analyzed by Western blotting. Equal amounts of lysates were fractionated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to Bio-Rad trans-blot nitrocellulose membranes. The nitrocellulose membrane was then incubated in blocking buffer (Tris-buffered saline containing 0.1% polysorbate 20 and 5% nonfat milk) for 1 h at room temperature. Then the membrane was incubated with the appropriate primary antibody (anti-MDM2 or anti-p53, 1:1,000 dilution) overnight at 4°C with gentle shaking. The membrane was then washed 3 times (15 min each) with washing buffer (Tris-buffered saline containing 0.1% polysorbate 20) and incubated with a goat antimouse antibody (1:3,000 dilution) for 1 h at room temperature. After washing 3 times with washing buffer, the protein band was detected and analyzed with a gel imaging system.
Biodistribution Studies
BALB/c mice (female; mean weight ± SD, 20 ± 2 g; age, 3–4 wk) were used in the study. The mice were inoculated with 1 × 107 MCF-7 cells in the right hind limbs, and the tumors were allowed to grow to a diameter of 1–2 cm. The mice were maintained using a standard diet, bedding, and environment, with free access to food and drinking water.
Twenty-four BALB/c mice with MCF-7 xenografts were randomly divided into 8 groups of 3 mice each (4 groups for injection of antisense probe, another 4 groups for injection of mismatch probe). Antisense probe or mismatch probe (7.4 MBq) was injected into each mouse via a tail vein. At 1, 2, 4, and 6 h after injection, the mice were sacrificed by cervical dislocation. Then the mice were dissected; tissues of interest (blood, heart, lung, liver, spleen, kidney, small intestine, skeletal muscle, thyroid, and tumor) were washed clean with cool physiologic saline and weighed, and their radioactivity was measured using a γ-well counter. Radioactivity results were recorded as the percentage injected activity per gram of tissue (%ID/g) and the ratio of tumor to skeletal muscle (T/M).
Molecular Imaging
Ten BALB/c mice with MCF-7 xenografts were randomly divided into 2 groups of 5 mice each (one group for injection of antisense probe, the other for injection of mismatch probe). Antisense probe or mismatch probe (7.4 MBq) was injected into each mouse via a tail vein. At 1, 4, and 10 h after injection, imaging was performed using a SPECT scanner (Millennium VG, Hawkeye; GE Healthcare) equipped with a low-energy, high-resolution, parallel-hole collimator. Static images (100,000 counts), obtained with a zoom factor of 2.0, were digitally stored in a 256 × 256 matrix. The ratio of T/M was calculated over the region of interest.
Statistical Analysis
The variables are expressed as average ± SD. The variables of RT-PCR and Western blotting were analyzed using variance analysis and least-significant difference t test, and other statistical comparisons of average values were performed with the Student t test. A P value less than 0.05 was considered significant.
RESULTS
Oligonucleotide Conjugation and Radiolabeling
The labeling efficiency of 99mTc-HYNIC-ASON and 99mTc-HYNIC-ASONM was 57.2% ± 2.98% (n = 5) and 56.3% ± 3.01% (n = 5), respectively; the specific activity was 1,450 ± 60.2 kBq/μg and 1,370 ± 55.4 kBq/μg, respectively; and the radiochemical purity for both was above 95% after purification.
Molecular Hybridization Activity
After the 99mTc-HYNIC-ASON was incubated with the unlabeled sense oligonucleotide, the radioactivity peak appeared late (Fig. 1).
Radiochromatogram obtained by reversed-phase Sep-Pak C18 analysis of 99mTc-HYNIC-ASON before (A) and after (B) 60 min of incubation with its sense complement.
RT-PCR
A gradual decrease of MDM2 mRNA expression was observed in cells treated with increasing antisense probe doses. Compared with the antisense probe, the mismatch probe had no effect on MDM2 mRNA expression levels. There were significant differences in MDM2 mRNA expression between the 100 nmol/L antisense probe group and the control group of 10% fetal bovine serum and between the 500 nmol/L antisense probe group and the control group of 10% fetal bovine serum (P < 0.01, n = 5). The p53 mRNA expression was not obvious, and no significant difference between either of the 2 antisense probe groups and the control was observed (P > 0.05, n = 5) (Fig. 2).
Effects of antisense and mismatch probes on MDM2 (A) and p53 (B) mRNA levels in MCF-7 cells. PCR products were analyzed with variance analysis and least-significant difference t test. Lane M, DNA standard marker; lane 1, control group of 10% fetal bovine serum; lane 2, 100 nmol/L antisense probe groups; lane 3, 500 nmol/L antisense probe groups; lane 4, 500 nmol/L mismatch probe groups. *P < 0.01, for comparisons with control group.
Western Blotting
Like the MDM2 mRNA levels, MDM2 protein expression levels decreased gradually with increasing antisense probe doses. Unlike the p53 mRNA levels, however, the p53 protein expression levels increased gradually with increasing antisense probe doses. There were significant differences in both MDM2 and p53 protein expression between the 100 nmol/L antisense probe group and the control group and between the 500 nmol/L antisense probe group and the control group (P < 0.01, n = 5). The mismatch probe had no effect on MDM2 and p53 protein expression (Fig. 3).
Effects of antisense and mismatch probes on MDM2 (A) and p53 (B) protein levels in MCF-7 cells. Protein was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis, followed by Western blotting. Protein density was analyzed with variance analysis and least-significant difference t test. Lane 1, control group of 10% fetal bovine serum; lane 2, 100 nmol/L antisense probe groups; lane 3, 500 nmol/L antisense probe groups; lane 4, 500 nmol/L mismatch probe groups. *P < 0.01, for comparisons with control group.
Biodistribution Studies
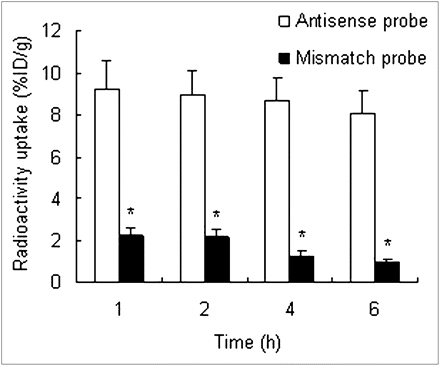
Biodistribution data are shown in Tables 1 and 2. At the various times after injection of the antisense and mismatch probes, these 2 probes had similar biodistributions, except for radioactivity uptake in the tumor. In the tumor, the mismatch probe concentration was significantly lower than the antisense probe concentration (P < 0.01, n = 3) (Fig. 4). Because of blood clearance, tumor uptake of the 2 probes gradually decreased. However, tumor uptake of the antisense probe still remained at a relatively high level until 6 h after injection. Between 1 and 6 h after injection of the antisense probe, its uptake ratio in T/M increased from 2.28 to 12.08.
Biodistribution of Antisense Probe in Mice Bearing MCF-7 Xenografts
Biodistribution of Mismatch Probe in Mice Bearing MCF-7 Xenografts
Radioactivity uptake in tumors after injection of antisense and mismatch probes. At all time phases, tumor radioactivity uptake of antisense probe is significantly higher than that of mismatch probe. Radioactivity uptake was analyzed with Student t test. *P < 0.01.
The 2 probes accumulated primarily in the kidney and liver. The other organs (blood, heart, lung, spleen, small intestine, skeletal muscle, and thyroid) showed low uptake of the probes. The radioactivity gradually decreased in the kidney and liver over time, indicating that the probes were cleared primarily through the urinary and digestion systems. In addition, the biodistribution of the 2 probes was characterized by quick blood clearance, with 7.42 ± 1.01 %ID/g and 7.39 ± 1.07 %ID/g remaining at 1 h after injection and 2.24 ± 0.22 %ID/g and 2.15 ± 0.35 %ID/g remaining at 4 h after injection.
Molecular Imaging
In nude mice bearing MCF-7 xenografts, the tumors were clearly imaged at 1, 4, and 10 h after injection of antisense probe. However, the tumors were not apparent on SPECT images at any time after injection of the mismatch probe. The ratio of T/M is significantly higher in the antisense groups than in the mismatch groups (Fig. 5). As predicted from the biodistribution studies, the liver and bladder were clearly imaged in both the antisense group and the mismatch group.
Imaging of MDM2 expression in MCF-7 xenografts in BALB/c mice using antisense (A) and mismatch (B) probes. Tumor (arrow) is clearly visualized after injection of antisense probe but is not clearly evident after injection of mismatch probe. At all time phases, ratio of T/M is significantly higher in antisense groups than in mismatch groups (C). Ratio of T/M was analyzed with Student t test. *P < 0.01.
DISCUSSION
Antisense imaging, a specific and noninvasive technology, is based on the complementarity of the constructs to the appropriate target mRNA. Pioneering work in this field was reported by Dewanjee et al. in 1994, who used a 15-mer phosphorothioate antisense oligodeoxyribonucleotide labeled with 111In to image constitutive c-myc gene expression in a murine mammary carcinoma model (21). Up to now, ASONs directed toward c-myc, bcl-2, CCND1, mdr1, hTERT, and other targets have been chosen for labeling with radionuclides (22–29), and the results of all these studies have facilitated the development of antisense imaging.
The design of an efficient ASON targeting MDM2 mRNA is critical. One of the limiting factors is the accessibility of the target RNA sequence because stable secondary structures might prevent ASON hybridization. Recently, the experimental and theoretic approach increased the probability of finding effective antisense molecules (30). Another key factor is the optimum length that the ASON needs to bind to its target. According to statistical calculations, the optimum size should be between 15 and 20 bases for a given oligonucleotide. According to the principles of ASON design and a previous study (31), the 20-mer ASON of 5′-GATCACTCCCACCTTCAAGG-3′ was selected for this study. Otherwise, considering the ASON stability in vivo, we used uniformly phosphorothioate-modified oligonucleotide.
Because the marker is in the process of chemical reaction and the radiolabeling product can change the ability of the marker to bind to the complementary chain, it is necessary to study the binding capacity after radiolabeling. The Sep-Pak C18 column separation principle is based on the material polarity difference. After the 99mTc-HYNIC-ASON is incubated with the unlabeled sense oligonucleotide, aliquots are removed for reversed-phase Sep-Pak C18 column chromatography analysis. Compared with proincubation, in radiolabeling the radiochemical peak appears late, indicating that 99mTc-HYNIC-ASON binds to the sense oligonucleotide for forming polarity that is molecularly lower. The ASON radiolabeled with 99mTc does not affect the combination capability with the sense oligonucleotide.
Although antisense probes can bind to the sense chain, we need to understand the cellular level, at which more complicated and environmental factors have additional influence regarding whether MDM2 mRNA ASON can specifically bind to the whole chain of MDM2 mRNA. For these reasons, we applied different concentrations of radiolabeled antisense and mismatch probes to transfect MCF-7 cells and detected MDM2 mRNA expression levels and the corresponding protein expression. Further, MDM2 and p53 form a negative-feedback loop; therefore, p53 gene expression also can reflect MDM2 gene expression. The tests also detected expression of the p53 gene. We examined antisense probe inhibitory action in the MCF-7 cell line and MDM2 and p53 gene expression.
One of the key questions is how to maximize the transfection of antisense probe into tumor cells. Because there are no direct receptors in the cell membrane for oligonucleotides, it is difficult for the cell to take up the probe directly. Numerous methods have been developed for DNA transfer, such as electroporation, liposomes, and viral vectors, but the development of cationic liposomes has progressed the furthest. The reduced immunogenicity and safety of cationic liposome delivery systems, compared with electroporation and viral vectors, make cationic liposomes widely used as biocompatible transporters of foreign genetic DNA (32). Considering these factors, in our study we selected Lipofectamine 2000 as the delivery vehicle for the probes to produce a higher transfection ratio.
The gradual decreasing of MDM2 mRNA and protein expression suggested that radiolabeled ASON binds to the target genes and inhibits their expression. The p53 mRNA expression is not obviously, changed, and corresponding protein expression increases gradually. The result is similar to the findings of Zhang et al. (33), demonstrating that MDM2 regulates p53 expression mainly by degradation of its protein levels rather than inhibition of its transcription. This result further confirms that the antisense probe is binding to the target genes. To confirm the specificity of binding of antisense probe to the target gene, we synthesized a mismatch probe with 4 mismatch bases. The mismatch probe had no effect on MDM2 and p53 expression.
At 1–6 h after injection of the antisense probe, its biodistribution in mice bearing MCF-7 xenografts showed that it was quickly cleared in the main organs of the body, except in the tumor. The uptake rates of the liver (11.26 ± 1.97 %ID/g) and kidneys (13.76 ± 2.47 %ID/g) were slightly higher than that of the tumor (9.21 ± 1.37 %ID/g) at 1 h after injection. Because the oligonucleotide was primarily metabolized through the liver and kidneys, the antisense probe preferentially accumulated in the liver and kidneys. Over time, radioactivity gradually decreased in the kidneys and liver. At 6 h after injection, the radioactivity was obviously lower in the kidneys and liver than in the tumor. Compared with the antisense probe, the mismatch probe had similar biodistribution, except in the tumor. The biodistribution results of these 2 probes could support the mechanism of antisense—that is, the specific hybridization of antisense probe to MDM2 mRNA results in the specific accumulation of radiolabeled ASON in the MDM2-expressing tumors.
The results of imaging of MDM2 expression in MCF-7 xenografts using the radiolabeled probes were consistent with the results of biodistribution. Tumors were clearly visualized after injection of the antisense probe but were not clearly imaged after injection of the mismatch probe. The results of imaging also confirmed the mechanism of the antisense probe. High retention in the liver and bladder is also an apparent obstacle for imaging the tumors in abdominal organs. However, this does not seem to be a problem for SPECT of MDM2 expression in breast carcinomas in our study, in which the tumor is anatomically well separated from the liver and bladder.
The results of our study prove that liposome-coated 99mTc-HYNIC-ASON targeting MDM2 mRNA can be used for the detection of MDM2 expression in breast carcinomas in vivo. Because MDM2 takes an important role in the occurrence, development, and treatment of many tumors, the next phase of our study is to investigate the possibility of using MDM2 antisense probe for imaging of MDM2 expression in more tumors.
CONCLUSION
The accumulation of liposome-coated 99mTc-labeled ASONs in breast cancer tissue is specific. The expression of MDM2, a critical oncogene, can be measured by liposome-coated 99mTc-HYNIC-ASON targeting MDM2 mRNA, which is a potential and promising candidate for the visualization of MDM2 expression in carcinomas.
Acknowledgments
We thank Prof. YongXue Zhang (Tongji Medical College, Huazhong University of Science and Technology, China) for his kind instructions during this study. We also thank the Department of Chemistry at Beijing Normal University for providing HYNIC for conjugation. This study was supported by Heilongjiang Province Office of Education (grant 11531192).
- © 2010 by Society of Nuclear Medicine
REFERENCES
- Received for publication April 6, 2010.
- Accepted for publication August 11, 2010.