Abstract
The majority of investigational studies of new diagnostic and therapeutic radiopharmaceuticals use murine animal models for preclinical assessments of pharmacokinetics and organ radiation dosimetry. Although mice and rats are widely available and relatively inexpensive, their smaller organ anatomy relative to that of humans can lead to considerable differences in organ dosimetry, thus complicating extrapolations of dose–response relationships to human patients. Nonhuman primates circumvent these problems in many respects but are increasingly becoming expensive and limited because of ethical considerations. With the recent completion of the dog genome project and the recognition of many similarities between canine and human cancers, dogs are increasingly being considered in cancer research and drug development. The main objective of this study was to construct a 3-dimensional computational phantom of a large dog on the basis of whole-body multislice CT data. Methods: A female hound cross underwent whole-body contrast-enhanced CT at a 2-mm slice thickness. On completion of the scan, the dog was euthanized, and the entire skeleton was harvested for a subsequent microCT investigation. The CT data were imported into a computational software program and used to create a polygon-mesh phantom of the entire animal. All of the major organs and bones were semiautomatically segmented and tagged to the CT slices. The phantom data were imported into a second software program and transformed to a nonuniform rational basis-spline surface phantom, allowing easy alteration of the phantom to simulate dogs of smaller or larger statures. A voxel-based version of the canine phantom was created by use of an in-house routine for subsequent import into the EGSnrc radiation transport code for photon and β-particle organ dosimetry. Results: The resulting voxel-based version of the canine phantom had a total body mass of 26.0 kg and a total body tissue mass (exclusive of wall organ content) of 24.5 kg. Although this University of Florida (UF) canine phantom displayed a total body mass intermediate between those of the Oak Ridge National Laboratory (ORNL) 5-y and 10-y stylized human phantoms of the MIRDOSE and OLINDA software codes, considerable differences were noted in organ photon cross-doses. For example, ratios of the specific absorbed fraction Φ(lungs ← liver)UF Dog to Φ(lungs ← liver)ORNL 5-y ranged from ∼30 at 10 keV to ∼3.5 at 1 MeV. Corresponding ratios of Φ(lungs ← liver)UF Dog to Φ(lungs ← liver)ORNL 10-y ranged from ∼6 at 10 keV to ∼1.3 at 1 MeV. Conversely, values of Φ(kidneys ← spleen) and Φ(liver ← spleen) were noted to be much lower (factors of 2–4) and much higher (factors of 2–15), respectively, in the canine phantom than in the ORNL human phantoms. These differences were attributed more to organ shape and position within the torso than to organ mass, because many of the canine organs closely approximated their counterparts volumetrically in the stylized pediatric human phantoms. Conclusion: The use of canine models, particularly in spontaneously occurring malignancies such as osteosarcoma, for preclinical testing of antineoplastic agents offers significant advantages over current murine models. However, the development of canine-specific technology is critical to the optimization of these studies. The UF canine dosimetry phantom described here aims to solve problems that could stem from the use of current human dosimetry models during radiopharmaceutical research.
With the recent completion of the canine genome project (1,2) and the recognition of the similarities between canine cancers and human cancers (3), it is now clear that dogs are an effective model for the preclinical development of novel antineoplastic agents (4). Rodents and larger animals, such as nonhuman primates, have been used in cancer research for many years. Although the availability and short life span of mice and rats make them useful investigative tools for new drugs, the results obtained from these studies must be extrapolated to humans, with possibly large uncertainties. In radiopharmaceutical research, the much smaller anatomic dimensions in rodents relative to those in humans can result in vastly different patterns of energy deposition among tissues of interest (5–8). As a result, dosimetry estimates and dose–response relationships are complicated and not easily extrapolated to humans (9). On the other hand, research performed with nonhuman primates can be more fruitful, but it is hardly the best option because of the limited availability and cost of these animals. Accordingly, cancer research with canine subjects is ideal because a large population is available [approximately 4 million dogs are diagnosed with cancer each year in the United States (3)] and many histologic and biochemical similarities exist between canine and human diseases (10,11).
Nevertheless, before canine radiopharmaceutical studies can reach their full potential, computational and dosimetry tools must be made available for researchers to fully use canine models of human disease. At present, no canine dosimetry model exists for evaluating radiation doses during preclinical studies. Veterinary doctors and researchers accordingly have had to rely on human dosimetry software, such as MIRDOSE3.1 (12) and OLINDA/EXM, version 1.0 (13), to perform their calculations (14–17). Although efforts are currently under way to update the OLINDA code to include voxel-based anatomic phantom dosimetry (18), present versions of both of these codes use stylized human phantoms from the Oak Ridge National Laboratory (ORNL) series (19).
The objectives of this study were to create a realistic anatomic phantom of an average dog and to use that model to establish an extensive database of photon and electron dosimetry for subsequent preclinical canine studies in molecular radiotherapy. In the resulting University of Florida (UF) hybrid canine phantom, nonuniform rational B-spline (NURBS) surfaces are used to model internal organ structure and outer body dimensions; therefore, this UF phantom takes advantage of both the scalability of existing stylized dosimetry phantoms and the anatomic realism offered in more modern image-based voxel phantoms. These techniques are similar to those used with the NURBS-based cardiac–torso phantom in nuclear medicine imaging studies (20–23).
MATERIALS AND METHODS
Animal Selection
This study was approved by the University of Florida Institutional Animal Care and Use Committee and was performed in accordance with the Institute for Laboratory Animal Research Guide for the Care and Use of Laboratory Animals. A 24-kg, 3-y-old female hound cross was selected on the basis of her size and breed but was deemed to be representative of a broad range of canine subjects and thus offered the external and internal anatomy necessary to establish an initial reference canine phantom. Before enrollment, the dog was certified to be disease free and not on any medications. Furthermore, the dog represented a phenotype that is commonly used as a model for human disease, including various forms of bone cancer (4).
CT
Anesthesia was induced intravenously with propofol infusion (120 mg) and maintained with 1%–2% isoflurane. The dog was scanned in a prone position by use of a Philips Tomoscan-M multislice CT scanner. The x-ray tube was set at 120 kVp and 10 mA. The field of view was 35 cm × 35 cm, and image reconstruction was performed with a grid of 512 × 512 pixels. The slice thickness was set at 2 mm. The scan was performed in 3 overlapping sections: head and neck, thorax and abdomen, and pelvis and tail. At the start of the study, the dog was given 200 mL of iohexol (300 mg/mL) for contrast enhancement, and this step was repeated with each scan section. On completion of the CT study, the dog was euthanized (20 mL of Beuthanasia-D Special; Schering-Plough Animal Health Corp.), and the complete skeleton was harvested. The individual bones were cleaned by use of an atraumatic technique (the bones were placed in a beetle box, and the insects were permitted to clean excess soft tissues on the exterior bone surfaces) and preserved for later ex vivo CT and in vitro microCT studies of the skeletal architecture at both macroscopic and microscopic levels.
Polygon-Mesh Model
A total of 29 tissues and 19 skeletal sites were selected for model construction of the canine anatomic phantom. As shown in Supplemental Table 1 (supplemental materials are available online only at http://jnm.snmjournals.org), these organs were primarily those listed in International Commission on Radiological Protection Publication 89 (24) for human radiologic protection dosimetry. All nonsegmented regions of the body, such as adipose tissue, skeletal muscle, major blood vessels, lymphatic tissue, and connective tissue, were denoted as residual soft tissues and were assigned as a single tissue region.
After CT, the resultant images were imported into 3D-Doctor (Able Software Corp.), a 3-dimensional (3D) modeling and image-processing software for tomography data. With this software, organs of interest were segmented semiautomatically within each axial CT image. Tissue regions were tagged with organ identifiers for later use in radiation transport modeling and 3D image visualization. The individual organ contours were subsequently joined in the assembly of a polygon-mesh model of each organ as well as the outer body contour of the animal. An example of this process for the canine lungs is shown in Supplemental Figure 1.
Some smaller organs within the CT images were difficult to visualize because of low image contrast. These included the pituitary gland, thymus, adrenal glands, and ovaries. These organs were inserted manually within the phantom by use of an anatomic text reference (25) and by concurrence with a veterinary oncologist.
NURBS Model
After construction within 3D-Doctor, organ-specific polygon-mesh models were imported into a second software program, Rhinoceros (McNeel North America), used to fit NURBS surfaces to polygon-mesh representations of the tissues and organs in the phantom. For manipulation of polygon-mesh anatomic models independently and effectively, Wavefront Object files were generated from 3D-Doctor in 5 different groups: the exterior body contour, the alimentary system, the respiratory system, other soft-tissue organs, and the skeleton. The groups of organs were then imported as different layers into Rhinoceros. In this manner, each layer could be turned on or off with objects within other layers being unaffected. Smooth NURBS surfaces were created from these polygon-mesh models on an organ-by-organ basis. First, several contours were obtained from the polygon-mesh models as needed, and NURBS surfaces were fit to these contours by use of tools within the software. NURBS surfaces were generated for all internal organs and tissues. Supplemental Figure 2 shows this process for the canine lungs. The single exception, however, was the skeleton, for which the complex 3D structure was best preserved within the polygon-mesh model generated from 3D-Doctor.
The small intestine and colon were difficult to visualize within the CT images, again because of low image contrast as well as their complex anatomic geometry. Consequently, these organ structures were created as follows. First, the outer boundaries of the canine gut were traced within the CT images by use of 3D-Doctor. Next, the CT images were used to define a central trace of the colon lumen, after which a NURBS pipe model was constructed to define the colon wall. A similar approach was used for the small intestine, with the small intestine length and wall thickness being matched to values indicated as “representative” in a canine anatomic text reference (25). The creation of the intestine and colon model of the UF canine phantom is shown in Supplemental Figure 3.
Several walled organs were modeled directly within Rhinoceros. These included the esophagus, heart, stomach, gallbladder, and urinary bladder. Visualization of the esophagus was possible in only some of the axial CT slices; therefore, as with the gut model, a NURBS pipe structure providing representations of the thickness and length of the esophagus that were consistent with both the CT images and the anatomic text reference was used (25).
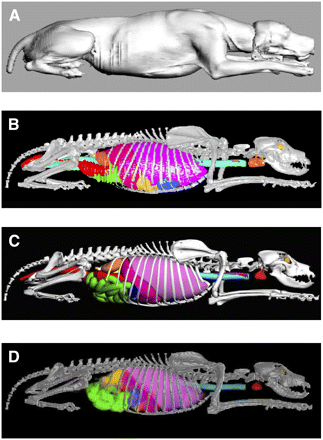
In the format of the polygon-mesh model, the ribs had a stair-step appearance because of the limited slice thickness of the organ CT image set and the 3D orientation relative to the slice plane. Consequently, the entire rib cage was remodeled with NURBS surfaces. As shown in Supplemental Figure 4, costal cartilage that was not fully represented in the polygon-mesh model was manually included in the NURBS surface version of the skeletal site. After model completion, it was seen that the urinary bladder of the subject was overly full because of the injection of the contrast agent at the time of CT. The urinary bladder was thus modified in Rhinoceros to reduce the organ to a more standard size, as shown on the right side of Supplemental Figure 5. A side-by-side comparison of the polygon-mesh version and the NURBS surface version of the UF canine phantom is shown in Figures 1B and 1C; a surface rendering of the original CT image set is shown in Figure 1A.
Visual comparison of different versions of UF hybrid canine phantom. (A) Surface rendering of original CT image set. (B) Polygon-mesh model generated from 3D-Doctor. (C) NURBS surface model generated from Rhinoceros. (D) Voxel-based model generated from MATLAB-based code Voxelizer.
Voxel Model and Radiation Transport Simulations
Internal dosimetry calculations were performed by use of the EGSnrc radiation transport code (26), which required a voxel-based version of the UF canine phantom. The phantom was prepared for voxel creation by export of the finalized NURBS-based phantom as a group of polygon-mesh objects, with each organ being assigned a relative level of importance, a step required for resolving organ overlap issues during the voxel creation process. The voxelized model was accomplished by use of an in-house MATLAB (MathWorks)–based code, Voxelizer (27). The resulting voxel-based version of the UF canine phantom is shown in Figure 1D. The final voxel resolution of the phantom was set at 2 × 2 × 2 mm3 primarily on the basis of the need to properly represent the epidermis or dermis of the animal as a single-voxel layer in the final model.
Four tissue media were used for the radiation transport calculations: air, soft tissue, skeleton (homogeneous mixture of bone and marrow), and lung (homogeneous mixture of soft tissue and air). Because there are no comprehensive reference values for tissues and organs within adult dogs, the elemental compositions and mass densities of these materials were taken to be those used in ORNL/TM-8381 for the ORNL stylized human phantom series (19). Photon and electron source energies of 10 keV–4 MeV were also chosen for radiation transport simulation at an energy grid equivalent to that used in ORNL/TM-8381. Particle histories were set at 108 particles per source organ at energies below 100 keV and 107 particles at energies above 100 keV.
Database of Electron- and Photon-Specific Absorbed Fractions
For all tissues and organs, the radiation transport results were used to calculate absorbed fractions (ϕ) and specific absorbed fractions (Φ) for the canine phantom. Supplemental Appendix A shows absorbed fractions and specific absorbed fractions for 36 photon source tissues and 30 target tissues. Supplemental Appendix B shows absorbed fractions and specific absorbed fractions for these same tissues for electron sources. Microsoft Excel versions of the appendices are available via e-mail request to the corresponding author. The dosimetry database includes the explicit consideration of paired organs (e.g., left and right lungs are independently considered as both source and target tissues) and the consideration of wall uptake of radiopharmaceuticals in alimentary tract organs (e.g., small intestine and colon walls).
Graphical inspection was performed for the specific absorbed fraction as a function of photon energy for all possible source and target tissue combinations (36 source regions × 30 target tissues, or 1,080 plots, as shown in Supplemental Appendix A). When the target tissue was found to be far from the source region (e.g., brain irradiated by a liver source) or was small in total volume (e.g., thyroid), coefficients of variation for the absorbed fraction increasingly exceeded 30%–50% at low photon energies (10–20 keV); at some point, no photon energy deposition events were recorded. In these situations, extrapolations based on log–log regression fits of low-energy photon specific absorbed fractions were performed by use of SigmaPlot, version 10 (Systat Software Inc.), to provide a complete and continuous energy-dependent dataset. Supplemental Table 2 shows the energy below which extrapolated data were generated. In some situations, statistical imprecision was noted at higher photon energies. In these situations, the reciprocity theorem was invoked, so that values of Φ(target ← source) (D: direct Monte Carlo) were approximated by corresponding values of Φ(source ← target) (C: converse Monte Carlo). In some situations, surrogate organs were assigned (e.g., the brain for the pituitary gland). In a manner similar to that implemented in ORNL/TM-3831 for stylized human phantoms (19), specific absorbed fractions below 10−10 kg−1 were set to zero. Although these techniques are approximate in nature, they are applied only for very low-energy photons when the specific absorbed fractions are orders of magnitude lower than the corresponding values for organ self-doses.
For the electron sources in Supplemental Table 3 and Supplemental Appendix B, values of absorbed fractions and specific absorbed fractions are tabulated for the source tissue and various adjacent tissues that receive bremsstrahlung photon irradiation and, at high energies, direct electron irradiation from emissions in the target tissue. Because bremsstrahlung absorbed fractions are subject to increasingly higher statistical errors as one moves further and further from the source tissue, an effective cutoff was applied; that is, no values of specific absorbed fractions were recorded if the values did not exceed 10−4 kg−1 at a source electron energy of 100 keV. With this rule in place, electron and bremsstrahlung contributions were still characterized for 13 different tissues in the vicinity of the liver, as shown in Supplemental Table 3. Values of absorbed fractions and specific absorbed fractions for electron sources in the liver were set to zero for all other target tissues.
RESULTS
The final organ volumes and organ masses as they appeared in the voxel-based version of the UF canine phantom (Fig. 1D) are shown in Supplemental Table 1. Tissues are listed by organ system, and the standardized ORNL/TM-8381 tissue densities assumed are noted as well. Residual soft tissues (e.g., tissues and organs not segmented) are given in the “additional tissues” category and include separable fat, muscle, major blood vessels, lymphatic nodes, and connective tissue. The recorded total body mass of the animal at the time of CT was 24 kg. The total body mass of the voxel-based version of the canine phantom was 26.0 kg, and with the removal of all interior fluids of walled organs, the total body tissue mass of the animal model was 24.5 kg.
The skeletal tissues in the UF canine phantom are homogeneous; therefore, we cannot at present report masses for cortical bone, trabecular bone, red bone marrow, and yellow bone marrow. The design of a more detailed skeletal model (including the trabecular and marrow cavity microstructures) for this phantom is currently in progress, with imaging by both ex vivo CT and spongiosa microCT. Consequently, the absorbed dose to the homogeneous skeleton was tabulated for both internal photon and internal electron sources. As new data on the skeletal microstructure and corresponding electron dosimetry become available, it will be possible to replace values for the homogeneous skeletal absorbed dose with values explicitly detailing energy deposition to the canine active bone marrow and skeletal endosteum.
Before the establishment of the UF canine phantom, the 10-y-old stylized human phantom of the MIRDOSE program was used as an approximate model for assessing internal organ doses in preclinical canine studies of both diagnostic and therapeutic radiopharmaceuticals. The selection of this particular humanlike phantom was based on considerations not only of total body mass but also of individual organ sizes and torso length. The total body mass of the ORNL 10-y human phantom is 33.2 kg (1.3 times heavier than the UF canine phantom), whereas the total body mass of the ORNL 5-y human phantom is 19.8 kg (80% of the mass of the UF canine phantom). To further explore the appropriateness of 5-y versus 10-y ORNL phantoms as surrogate canine phantoms, we present individual organ masses for all 3 phantoms in Supplemental Figure 6. Five of the organs in the UF canine phantom are significantly smaller than those in either ORNL human phantom: adrenal glands, brain, gallbladder wall, pancreas, and thyroid. The colon wall and skin are noted to be appreciably larger than their ORNL organ models. The total body mass, as well as the residual soft tissues, of the UF canine phantom is between those of the 2 ORNL human phantoms, whereas the total skeletal mass is more closely approximated by that of the 5-y human phantom.
DISCUSSION
Of importance to photon cross-fire are not only the masses of the source and target organs but also their 3D shapes, locations, and positions, which can vary considerably between image-based anatomic models, such as the UF canine phantom, and stylized anatomic models, such as the ORNL human phantoms. In Supplemental Figures 7A–7D, we compare selected values of specific absorbed fractions in various target tissues for photon sources localized in the skeleton, liver, spleen, and kidneys, respectively, within each of 3 phantoms: UF canine, ORNL 5-y-old, and ORNL 10-y-old phantoms. Although the total body mass of the UF canine phantom is between those of the ORNL 5-y and 10-y phantoms, the skeletal mass is very close to that of the ORNL 5-y phantom. The values of Φ(lungs ← skeleton) in the UF canine phantom are between the values of the 2 stylized human phantoms, whereas the values of Φ(small intestine wall ← skeleton) are considerably lower in the UF canine phantom (Supplemental Fig. 7A). The data in Supplemental Figure 7B for photon sources in the liver indicate substantially higher values of absorbed doses per photon emission to the lungs and stomach wall in the UF canine phantom than are indicated from transport simulations in the ORNL human phantoms.
For photon sources in the spleen (Supplemental Fig. 7C), the cross-doses to the kidneys in the UF canine phantom are much lower than those seen in either the ORNL 5-y or the ORNL 10-y phantom. In contrast, both ORNL human phantoms show much lower values of Φ(liver ← spleen) across all photon energies than are indicated by the internal organ anatomy of the UF canine phantom. In Supplemental Figure 7D, photon sources are localized in both kidneys, and the cross-doses to the small intestine wall and liver are explored as a function of photon energy and phantom anatomy. The values of Φ(small intestine wall ← kidneys) are higher in the UF canine phantom at energies below 100 keV, above which the values are best approximated by the smaller, ORNL 5-y phantom. Cross-organ irradiation of the liver by photon sources in the spleen are substantially lower in the UF canine phantom than in either ORNL human phantom.
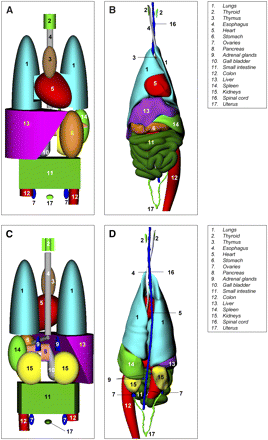
Differences in photon cross-fire may be attributed to several parameters, such as the masses of the source and target organs as well as their shapes, positions, and orientations with respective to one another in a given anatomic phantom. For example, consider the large discrepancies in Φ(lungs ← liver) and Φ(stomach ← liver) between the ORNL phantoms and the UF canine phantom (Supplemental Fig. 7B). Mass differences between the liver and the lungs in the ORNL 10-y phantom are very similar to those in the UF canine phantom (Supplemental Fig. 6); therefore, differences in photon cross-fire must be attributed to issues of 3D spatial orientation between organ pairs. Side-by-side views of the torso internal organ anatomy of the ORNL 10-y stylized human phantom and the UF canine phantom are shown in both anterior and posterior views in Figure 2. A large portion of the superior surface of the liver in the canine phantom is in direct contact with the inferior surfaces of the left and right lungs (Figs. 2B and 2D). Similarly, a sizable portion of the inferior surface of the canine liver is in direct contact with the latter reaches of the canine stomach (Fig. 2B). In contrast, Figures 2A and 2C show that virtually no direct organ-to-organ contact is maintained in the more fixed and rigid organ modeling of the ORNL 10-y human phantom. Consequently, photons at all energies have more tissue attenuation and scatter in the stylized phantom than in the more realistic dog anatomy.
Anatomic comparisons of ORNL 10-y stylized phantom (A and C) and UF canine phantom (B and D) in anterior view (A and B) and posterior view (C and D).
We have also made calculations of electron transport in the voxel-based version of the UF canine phantom across the energy range of 10 keV–4 MeV. Representative values of absorbed fractions for electron self-irradiation are shown in Supplemental Figure 8. The data show that for smaller and smaller organs and for higher and higher electron energies, the assumption of full energy absorption becomes increasingly unrealistic in the canine organ anatomy. Departures from full energy absorption become significant at approximately 300–400 keV in larger organs, such as the liver, spleen, and kidneys, and at much lower energies (∼100 keV) in smaller organs, such as the thyroid and pituitary gland.
CONCLUSION
The use of canine models, particularly for spontaneously occurring malignancies, such as osteosarcomas, in preclinical testing of antineoplastic agents offers significant advantages over the use of current murine models. However, the development of canine-specific technology is critical to the optimization of such studies. The UF canine dosimetry phantom described in this article aims to solve problems that could stem from the use of current human dosimetry models during radiopharmaceutical research. Future studies will further define the masses and skeletal site distributions of cortical bone, trabecular bone, red bone marrow, and yellow bone marrow in this same canine model on the basis of ex vivo CT and spongiosa microCT image analyses. Finally, the technique used, namely, NURBS surface modeling, can be extended to the construction of models of other dog breeds and animal sizes and can be used to alter body shape and rest-of-body mass to more explicitly model individual animals in preclinical dosimetry studies. This same technique is currently being used to expand the availability of human phantoms for internal dose assessments as well (18,28).
Acknowledgments
This study was supported in part by NCI grant F31 CA130165, by the National Cancer Institute, Shands Teaching Hospital Endowment for Oncology, and by the Razors 4 Raul Foundation for Research Toward a Cure for Osteogenic Sarcoma.
Footnotes
-
COPYRIGHT © 2008 by the Society of Nuclear Medicine, Inc.
References
- Received for publication August 25, 2007.
- Accepted for publication November 20, 2007.