Abstract
Earlier reports described the preferential uptake of d-amino acids in tumor-bearing mice. Moreover, it was shown that in tumor cells in vitro the l-amino acid transporter system seemed to lack stereospecificity. Because of the successful results with 123/125I-2-iodo-l-phenylalanine, 123/125I-2-iodo-d-phenylalanine was developed, and its tumor-detecting characteristics were evaluated in vivo. Methods: 123I labeling of 2-iodo-d-phenylalanine was performed with a kit formulation by use of Cu1+-assisted nucleophilic exchange. 123I-2-Iodo-d-phenylalanine was evaluated in R1M tumor–bearing athymic mice by dynamic planar imaging (DPI) and dissection. The in vivo stability of the tracer was tested by high-performance liquid chromatography. Tumor tracer retention and tracer contrast were evaluated as a function of time. Two-compartment blood modeling from DPI results and dosimetric calculations from biodistribution results were carried out. Moreover, 125I-2-iodo-d-phenylalanine and 18F-FDG uptake in acute inflammation was investigated. Results: 123I-2-Iodo-d-phenylalanine was metabolically stable. Fast, high, and specific tumor retention was observed. Two-compartment modeling confirmed the fast clearance of the tracer through the kidneys to the bladder, as observed by DPI and dissection. Moreover, compared with the l-isomer, 123I-2-iodo-d-phenylalanine demonstrated faster clearance and faster uptake in the peripheral compartment. No accumulation in the abdomen or in the brain was noted. Dosimetry revealed that 123I-2-iodo-d-phenylalanine demonstrated a low radiation burden comparable to those of 123I-2-iodo-l-phenylalanine and 123I-2-iodo-l-tyrosine. Although 123I-2-iodo-d-phenylalanine showed a tumor retention of only 4%, the tumor contrast was increased up to 350% at 19 h after injection. Conclusion: 123I-2-Iodo-d-phenylalanine is a promising tracer for diagnostic oncologic imaging because of its high, fast, and specific tumor uptake and fast clearance from blood.
At present, more and more interest is being shown in nuclear medicine–based tumor detection by PET and SPECT. The most prominent example is 18F-FDG, which is routinely used for oncologic imaging, but because of its high uptake in the brain and in inflamed tissues, new, more specific oncologic imaging tracers are required (1).
Both increased amino acid transport across the cell membrane and an increased rate of protein synthesis are early features of malignant transformation. Amino acid transport of types A and L has been shown to be increased in tumor cells relative to normal tissue, and these transport systems have been the major focus of the development of amino acid tracers for oncologic imaging (1,2).
Recently, various radiolabeled l-amino acids were successfully developed to overcome the limitations of 18F-FDG. Moreover, it was demonstrated that the membrane transport of the amino acids reflects the malignancy of the cells and that the incorporation of radiolabeled amino acids into cell proteins is not necessary. The increased amino acid uptake is directly related to the metabolic requirements of the tumor cells (1,3). 123I-3-Iodo-α-methyltyrosine is at present the only amino acid tracer routinely used for SPECT but, because of its marked long-term renal accumulation, the development of other radiolabeled amino acid tracers is necessary.
Tamemasa et al. (4) suggested the use of d-amino acids as specific tumor-detecting agents. They showed the preferential uptake of some unnatural radiolabeled d-amino acids in comparison to the l-isoforms in tumor-bearing mice. Moreover, Yanagida et al. (2) showed in tumor cells in vitro that system L seems to lack stereospecificity; d-amino acids are transported with a high affinity by l-amino acid transporter (LAT) system subtype 1 (LAT1). The latter, together with the slower excretion of d-amino acids from the tumor cells, could be an important rationale to develop d-amino acids as tumor diagnostic agents (5). Moreover, a lower radiation burden is expected from d-amino acids than from the l-isoforms because of their faster clearance from blood as a result of both negligible tissue distribution and incorporation into cell proteins.
Our research group developed 123/125I-2-iodo-d-phenylalanine and demonstrated its uptake in vitro in an R1M (rhabdomyosarcoma) cell model (6). The d-isomer was taken up by LAT1, was overexpressed in many tumors, and accumulated more slowly than, but in the same amounts as, 125I-2-iodo-l-phenylalanine. In this study, we evaluated 123/125I-2-iodo-d-phenylalanine as a potential new tumor diagnostic agent in vivo in an R1M athymic mouse model by means of dissection and dynamic planar imaging (DPI).
MATERIALS AND METHODS
All of the conventional products mentioned were at least of analytic or clinical grade and were obtained from Sigma-Aldrich. The solvents were of high-performance liquid chromatography (HPLC) quality and were obtained from Chemlab.
Synthesis of 2-Iodo-d-Phenylalanine Precursor
2-Iodo-d-phenylalanine was prepared from 2-bromo-d-phenylalanine (PepTech Corp.) by analogy with 2-iodo-l-phenylalanine as described previously (3). Briefly, Cu1+-assisted nucleophilic exchange under the same acidic and reducing conditions was used, and 2-iodo-d-phenylalanine and 2-bromo-d-phenylalanine were recovered separately by reversed-phase HPLC (RP-HPLC). Subsequently, 2-iodo-d-phenylalanine was obtained by evaporation of the mobile phase. Identification and quality control were achieved by liquid chromatography–mass spectroscopy, thin-layer chromatography, and RP-HPLC. Chiral chromatography was used to confirm the chiral purity (3).
Radiochemistry
123/125I-2-Iodo-d-Phenylalanine.
Radioiodination with 123I− (222 MBq; 10–20 μL) or 125I− (37 MBq; 10 μL) (Nordion Europe) of 1.0 mg of 2-iodo-d-phenylalanine was performed by Cu1+-assisted isotopic exchange under acidic and reducing conditions as described previously (3). The reaction mixture was rendered isotonic, the pH was adjusted to at least 4, and free 123I− was removed by Ag membrane filtration. Quality control was achieved by RP-HPLC with Sep-Pak C18 (Waters, Belgium), and chiral chromatography was used to confirm the chiral purity (3).
123I-Iodo-Human Serum Albumin.
Radioiodination of human serum albumin (HSA) with 123I (Nordion Europe) was performed by electrophilic substitution with the IODO-GEN (Pierce) technique as described previously (3).
In Vivo Experiments
Laboratory Animals.
The study protocol was approved by the local ethical committee for animal studies and was within the rules of Belgian legislation. Guidelines of the National Institutes of Health principles of laboratory animal care were followed.
Water and food were freely available during the experimental period.
Male Swiss nu/nu mice (n = 30) (Bioservices) were injected subcutaneously in the right flank (armpit region) with 5 × 106 R1M rhabdomyosarcoma cells. Normal tumor growth curves were obtained by use of sliding caliper measurements (3,7). For the inflammation model, male NMRI mice (n = 5) (in-house breeding program) were injected in the right biceps brachii with 25 μL of turpentine to create acute inflammation.
During all imaging experiments, the animals were anesthetized by intraperitoneal injection with 75 μL (1.5 mg) of a solution containing pentobarbital at 20 mg/mL (Nembutal [Ceva Santé Animale], 60 mg/mL). For the biodistribution experiments involving dissection, the animals were sacrificed by cervical dislocation without sedation, and the organs of interest were dissected.
123/125I-2-Iodo-d-phenylalanine, 18F-FDG, and 123I-iodo-HSA were injected intravenously in the lateral tail vein.
DPI.
Imaging was performed as described previously in the planar mode with a γ-camera (GCA-9300A/hg; Toshiba) equipped with a high-resolution parallel-hole collimator (3). All experiments were conducted with the same mouse population to limit the number of animals needed.
In order to perform semiquantitative analysis, a syringe with known 123I-2-iodo-d-phenylalanine activity (measured with a calibrated ionization chamber; Capintec CRC-15R [Ramsey]) was used for dose calibration. Syringes containing 123I-2-iodo-d-phenylalanine were counted with the same Capintec chamber before injection. Tracer uptake was expressed as the differential uptake ratio (DUR): (counts in tissue × pixels in total body)/(pixels in tissue × counts in total body). The latter calculation was performed in accordance with the conclusions and recommendations of Boellaard et al. (8) and Thie (9).
At first, a 123I-iodo-HSA study was performed to measure the relative blood-pool distribution to correct the uptake of 123I-2-iodo-d-phenylalanine in the studied organs for blood-pool activity (3).
DPI experiments were started when the tumor reached a volume of 1 cm3. Immediately after injection of 18.5 MBq of 123I-2-iodo-d-phenylalanine, a dynamic acquisition was performed; this acquisition was followed by a displacement study with l-phenylalanine (200 μL of a solution at 145 mmol/L intravenously) at steady state. Regions of interest (ROIs) were drawn by use of MRI maximum-intensity projection as described previously (3). 123I-2-Iodo-d-phenylalanine uptake by tumors was compared with uptake in the contralateral background area, and the ratio of tumor to contralateral background (RTB) was calculated. The significance of displacement of 123I-2-iodo-d-phenylalanine activity by l-phenylalanine was calculated for a 95% confidence interval.
To test the tumor retention of 123I-2-iodo-d-phenylalanine, 3 R1M tumor–bearing athymic mice were injected with 37 MBq of the tracer. At 1, 6.5, 16, 19, 21, 24, and 31 h after injection, a 15-min static image was acquired (1,024 × 1,024 matrix; field of view of 23.5 × 12.46 cm; photopeak window set at 15% around 159 keV). The tumor retention of the tracer was calculated relative to the time point 1 h after injection. The tumor contrast was defined as DUR relative to the 1-h time point: relative DUR = percent injected activity per tumor pixel divided by percent injected activity per total pixels at 1, 6.5, 16, 19, 21, 24, or 24 h; the result was divided by DUR at steady state (1 h).
Dissection Analysis of Biodistribution of 123I-2-Iodo-d-Phenylalanine in Tumor Model.
The injected dose was calculated as described previously (3). The same mouse population as that used for the DPI experiments was used for these experiments. Thirty R1M tumor–bearing athymic mice were injected with 7.4 kBq of 123I-2-iodo-d-phenylalanine 6 d after the last imaging experiment was performed. At various time points (2, 5, 10, 15, 30, 45, 60, 90, 120, and 180 min) after injection, 3 animals per time point were sacrificed. The amount of radioactivity in the organs and tissues was calculated as the differential absorption rate (DAR): (activity in tissue × total body weight)/(weight in tissue × activity in injected dose) (3).
Two-Compartment Modeling.
The data obtained by DPI and dissection for 123I-2-iodo-d-phenylalanine were fit to a 2-compartment model with intravenous bolus injection, without a lag time, and with first-order elimination by use of WinNonlin 4.0.1 (Pharsight Corp.) (Fig. 1). This kinetic model was chosen by analogy with 123I-3-iodo-α-methyltyrosine (10) The primary parameters V1 (apparent volume of distribution of the central compartment), k1,0 (velocity of elimination from the central compartment), k1,2 (velocity of distribution from the central compartment to the peripheral compartment), and k2,1 (velocity of distribution from the peripheral compartment to the central compartment) were determined. To obtain the primary parameters for 123I-2-iodo-l-phenylalanine, data from earlier work were used for the calculations (3).
Graphic presentation of 2-compartment model applied to blood DPI data for 123I-2-iodo-d-phenylalanine. IV = intravenous; V2 = apparent distribution volume for peripheral compartment.
Dissection Analysis of Biodistribution of 125I-2-Iodo-d-Phenylalanine in Inflammation Model.
Inflammation-bearing NMRI mice (n = 5) were injected with 7.4 kBq of 125I-2-iodo-d-phenylalanine together with 16 MBq of 18F-FDG at 24 h after turpentine injection. After 30 min, the mice were sacrificed, and the amount of radioactivity in the organs and tissues was calculated as the DAR (3).
Metabolism Study of In Vivo Stability of 125I-2-Iodo-d-Phenylalanine.
The blood collected in the dissection study was used for a metabolism study of 125I-2-iodo-d-phenylalanine as a function of time as described previously (3).
Dosimetric Calculations
Mean time–activity curves, expressed as percent injected activity per weight of tissue (%IA/weighttissue) or percent injected activity of tissue (%IAtissue), were generated for the organs of interest from the dissection experiments. Source organ residence times were determined from time integration of the biexponential fit to the experimental biodistribution data. Fitting was performed with the SPSS 12.0 software package (SPSS Inc.). In order to fit the experimental data, 2 extra time points were generated, at 792 min and 2,376 min after injection (1 and 3 times the half-life of 123I, respectively), by considering only the physical decay of the tracer and thus assuming the worst-case scenario. The latter procedure was performed because dissection was conducted only until 180 min after injection of the tracer. Extrapolation of the animal biodistribution to the reference human adult biodistribution was performed by assuming that either the %IA/weighttissue or the %IAtissue values were equal in mice and humans. Absorbed radiation dose estimates then were calculated for the target organs by applying MIRD methodology (11) for healthy adults with the MIRDOSE3.0 software package (MIRD Committee, Society of Nuclear Medicine, Reston, VA). The absorbed dose estimate for the urine bladder wall was calculated by use of the dynamic bladder model with a bladder voiding interval of 4.8 h as described by Cloutier et al. (12). The dosimetric calculations were performed not only on the data mentioned in this article but also on previously reported dissection data for 2-iodo-l-phenylalanine and 2-iodo-l-tyrosine to compare the new d-isomer tracer to other, newly developed amino acid analogs (3,13).
The parameters of the exponential fitted curves in SPSS were applied for hierarchical clustering of the organs by use of the average linkage algorithm. This analysis was performed to discover which organs exhibit the same behavior for 123I-2-iodo-d-phenylalanine tracer biodistribution.
RESULTS
Synthesis and Radiolabeling of 2-Iodo-d-Phenylalanine
Yields of up to 65% were obtained for the precursor synthesis of 2-iodo-d-phenylalanine from 2-bromo-d-phenylalanine by the Cu1+-assisted nucleophilic exchange method.
Radioiodination of 2-iodo-d-phenylalanine by Cu1+-assisted isotopic exchange, followed by Ag membrane filtration, resulted in a radiochemical purity of >99% and specific activities of 65 GBq/mmol (123I labeling) and 11 GBq/mmol (125I labeling).
In both cases, it was shown by chiral HPLC that there was no detectable amount of l-isomer analogs.
In Vivo Results
In Vivo Stability of 125I-2-Iodo-d-Phenylalanine.
HPLC analysis of mouse blood showed 4.8% free iodide at 90 min after injection. No other metabolites were detected. Ethylenediaminetetraacetic acid did not have any influence on the deiodination of 125I-2-iodo-d-phenylalanine.
DPI.
DPI with 123I-iodo-HSA revealed no significant difference between blood flow in the tumor and blood flow in the reference contralateral leg: RTB = 1.1 ± 0.2 (mean ± SD).
At equilibrium (plateau phase), net 123I-2-iodo-d-phenylalanine uptake in the tumor was high. An overall mean 123I-2-iodo-d-phenylalanine uptake by tumors of 1.00 ± 0.02 was obtained (Fig. 2). The administration of l-phenylalanine resulted in a significant displacement of the 123I-2-iodo-d-phenylalanine activity from the tumor; the radioactivity in the tumor showed a decrease of 9.6% ± 2.3% from the initial uptake value (P < 0.05) after the intravenous administration of l-phenylalanine.
Overall mean 123I-2-iodo-d-phenylalanine uptake (DUR) in tumor and contralateral background region as function of time, as determined by DPI (n = 30).
123I-2-Iodo-d-phenylalanine elimination from blood showed renal clearance to the bladder (Fig. 3). High tracer uptake was noted in the zone of the pancreas; it reached a peak value of 3.3 ± 0.2 at 2 min after injection but decreased rapidly (Fig. 4). Because of superposition of the pancreas with other organs, such as the left kidney and the stomach, the results of uptake in the pancreas are overestimated (14). Low uptake of radioactivity was observed in the thyroid (1.08 ± 0.06) and in the brain (0.76 ± 0.05) (Fig. 4).
Uptake of 123I-2-iodo-d-phenylalanine (DUR) as function of time, as determined by DPI, and clearance of tracer through kidneys to bladder. Kinetic fitting of observed blood curve for 123I-2-iodo-d-phenylalanine tracer distribution with theoretic curve (solid line) was done with 2-compartment model (n = 30).
Uptake of 123I-2-iodo-d-phenylalanine (DUR) in pancreatic zone, brain, and thyroid (n = 30).
The retention study for 123I-2-iodo-d-phenylalanine showed that only 4% of the initial 123I-2-iodo-d-phenylalanine uptake remained in the tumor at 31 h after injection. However, when tumor contrast was evaluated, an increase of 352% at 19 h after injection was observed (Table 1).
Retention of 123I-2-Iodo-d-Phenylalanine Activity in Tumors, as Determined by Planar γ-Scintigraphy (n = 3)
Dissection Analysis of Biodistribution.
Biodistribution data for 125I-2-iodo-d-phenylalanine at 2, 30, 60, and 120 min after injection in R1M tumor–bearing athymic mice are shown in Table 2. Those for 125I-2-iodo-l-phenylalanine were reported previously (3).
Biodistribution Analysis by Dissection of 125I-2-Iodo-d-Phenylalanine in R1M Tumor–Bearing Athymic Mice (n = 3)
The net 125I-2-iodo-d-phenylalanine uptake by tumors reached equilibrium at 30 min with a mean ± SD DAR of 3.9 ± 1.2. At the same time point, 125I-2-iodo-d-phenylalanine activity in blood and in the contralateral leg tissue reached DARs of 1.3 ± 0.2 and 1.1 ± 0.2, respectively.
125I-2-Iodo-d-phenylalanine was cleared very fast through the kidneys, without accumulation. High uptake of radioactivity was observed in the pancreas. Very low accumulation of radioactivity was observed in other abdominal organs, such as the liver, small intestine, and large intestine, the lungs, or the brain.
Comparison of 18F-FDG uptake with 125I-2-iodo-d-phenylalanine uptake at the inflammation site resulted in ratios for inflamed muscle to contralateral muscle of 10.5 ± 2.1 and 1.43 ± 0.09, respectively.
Two-Compartment Modeling.
The results obtained by 2-compartment modeling are shown in Figure 4 and Table 3. They showed that 123I-2-iodo-d-phenylalanine biodistribution analyzed by DPI and dissection fitted the theoretic curve for the proposed kinetic model (all R2 values were >0.95). In comparison with its l-isomer, 123I-2-iodo-d-phenylalanine was cleared from blood almost 2 times faster. Moreover, 123I-2-iodo-d-phenylalanine was taken up in the peripheral compartment faster, but its velocity of elimination from this compartment was comparable to that of 123I-2-iodo-l-phenylalanine. Both biodistribution methods showed the same kinetic patterns.
Kinetic Parameters of 2-Compartment Model Applied to Blood DPI Data for 123I-2-Iodo-d-Phenylalanine and 123I-2-Iodo-l-Phenylalanine
Dosimetry
Mean radiation dose estimates were calculated for the reference human adult by use of time–activity curves and organ residence times derived from mouse biodistribution data. The MIRDOSE3.0 program was applied, assuming that the values for either the percent injected activity per gram (%IA/g) of tissue or the percent injected activity (%IA) for 123I-2-iodo-d-phenylalanine, 123I-2-iodo-l-phenylalanine, and 123I-2-iodo-l-tyrosine in mice are equal to those in humans. Dosimetry results obtained from both estimates are summarized in Table 4. They show that the highest equivalent organ doses were reached for the pancreas and the stomach. For selection of the highest equivalent organ dose for each organ by both dosimetry estimates, this worst-case scenario would result in effective doses of 0.0330, 0.0375, and 0.0542 mSv/MBq for 123I-2-iodo-d-phenylalanine, 123I-2-iodo-l-phenylalanine, and 123I-2-iodo-l-tyrosine, respectively. Although no statistically significant difference was observed between the 2 phenylalanine analogs (P = 0.378), the tyrosine analog differed significantly from the other tracers (P = 0.041).
Dosimetric Calculations and Estimation of Worst-Case Effective Doses for 123I-2-Iodo-d-Phenylalanine, 123I-2-Iodo-l-Phenylalanine, and 123I-2-Iodo-l-Tyrosine (n = 3)*
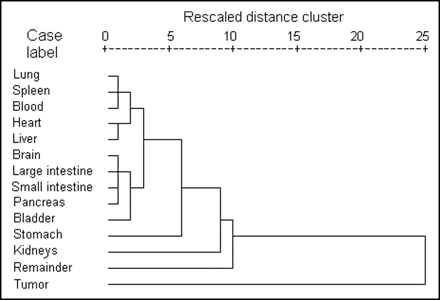
The clustering results for the coefficients of the exponential models used in the dosimetric calculations are visually presented by the dendrogram in Figure 5. It shows the closeness of the different organs with respect to 123I-2-iodo-d-phenylalanine pharmacokinetic behavior. The dendrograms for 123I-2-iodo-l-phenylalanine and 123I-2-iodo-l-tyrosine are not shown, but they demonstrated the same behavior. It is clear that the tumor, the remainder of the body, the kidneys, and the stomach are separate groups, with 2 additional clusters: blood group (blood, lungs, spleen, heart, and liver) and other-organ group (brain, large and small intestines, pancreas, and bladder). When the elimination coefficients only are examined (dendrogram not shown), an identical, even more pronounced, pattern was observed, with the exception of the stomach, which belonged to the blood group. Indeed, the kinetics of elimination from the stomach were similar to those for the blood group organs which, however, did not show uptake kinetics under the experimental conditions applied. These clustering results indicate that not every organ should be considered separately in full pharmacokinetic modeling because of similar pharmacokinetic behaviors within the groups.
Hierarchical clustering with average linkage algorithm for exponential curve parameters obtained by nonlinear regression of dissection data for 2-iodo-d-phenylalanine.
DISCUSSION
Tamemasa et al. reported the preferential uptake of d-amino acids in tumor-bearing mice (4,5). Moreover, Yanagida et al. showed in tumor cells in vitro that the LAT transporter seems to lack stereospecificity; d-amino acids are transported with a high affinity by LAT1 (2). Because of the successful results obtained with 125I-2-iodo-l-phenylalanine, 125I-2-iodo-d-phenylalanine was developed, and its tumor-detecting characteristics were evaluated.
In vitro in R1M cells, the Michaelis–Menten constant (Km) values for 125I-2-iodo-d-phenylalanine, 125I-2-iodo-l-phenylalanine, and the natural amino acid l-phenylalanine were similar. All compounds were transported by the same obligatory exchange system, LAT1 (6), confirming the hypothesis made by Yanagida et al. (2). In vitro (full minimal essential medium buffer), the accumulation of 125I-2-iodo-d-phenylalanine in R1M cells over a longer period of time (up to 24 h) was noted, although no incorporation into cell proteins was found, just as with the l-isomer (2,15).
Given the above-mentioned characteristics, the new tracer 123I-2-iodo-d-phenylalanine was evaluated in vivo for its potential application for oncologic diagnostic imaging by DPI.
Like 2-iodo-l-phenylalanine, the d-isomer could be synthesized easily and radioiodinated quantitatively with a kit formulation by use of Cu1+-assisted nucleophilic exchange (3). The same precursor yields and the same tracer specific activities were obtained for both tracers. No metabolism of 125I-2-iodo-d-phenylalanine took place. Only minor dehalogenation was observed; this dehalogenation was somewhat lower that that of the l-isomer and was reflected by the low radioactivity uptake in the thyroid. Thus, oral administration of stable potassium iodide before injection of 123I-2-iodo-d-phenylalanine should protect the thyroid from radioiodine poisoning.
Concerning the DPI data for 123I-2-iodo-d-phenylalanine, tracer kinetics were in accordance with the proposed 2-compartment model. Moreover, because of its higher k1,0 value and thus faster elimination from blood, 2-iodo-d-phenylalanine will demonstrate better tumor contrast at the same time point than its l-isomer. Additionally, 2-iodo-d-phenylalanine was taken up by the peripheral compartment faster than 2-iodo-l-phenylalanine (k1,2 for the d-isomer ≈ 1.4 times k1,2 for the l-isomer), but both tracers were eliminated at equal velocities from the peripheral compartment (k2,1 for the d-isomer ≈ k2,1 for the l-isomer). These findings, together with the very fast 123I-2-iodo-d-phenylalanine washout from the pancreas, will lead to faster and higher tumor uptake and thus better tumor contrast for 123I-2-iodo-d-phenylalanine than for the l-analog. This hypothesis was confirmed by the results of the 123I-2-iodo-d-phenylalanine tumor retention study. Although only 4% of the initial 123I-2-iodo-d-phenylalanine uptake remained in the tumor after 31 h, the tumor contrast became much more favorable for oncologic imaging than that obtained with 123I-2-iodo-l-phenylalanine, with a retention of 11%; whereas the latter showed a decrease in tumor contrast, 123I-2-iodo-d-phenylalanine showed an increase in contrast of 352% at 19 h after injection (Table 1). These results demonstrate the capability of 2-iodo-d-phenylalanine as a tumor-detecting agent in SPECT and indicate the potential of 2-iodo-d-phenylalanine as a radiotherapeutic agent. Indeed, metaiodobenzylguanidine, which showed tumor uptake of only 2% of the injected dose, has become an effective antineuroblastoma agent, in addition to its diagnostic tumor imaging application. Apart from its tumor-targeting properties, the new tracer 2-iodo-d-phenylalanine is metabolically stable but shows lower liver uptake and accumulation than metaiodobenzylguanidine and is rapidly cleared from the circulation by the kidneys; these properties are ideal starting points for systemic radiotherapy (16–18).
Different absolute values were obtained when data from DPI and data from dissection were used, but the same kinetic trends were observed. Indeed, this finding is emphasized by the ratio 123I-2-iodo-d-phenylalanine to its l-isomer in Figure 1.
Although the kinetic modeling of both tracers showed similar trends for DPI and dissection, the values differed significantly from each other. This finding could be explained by the difference in data acquisition; the ROI for blood in DPI also includes the heart muscle. Although the dissection results showed that no accumulation of the tracer took place in the heart muscle (primary energy source: glucose), the latter tissue could have an additional effect on the total counts in the ROI, resulting in higher values.
Regarding tumor uptake, DPI showed that 123I-2-iodo-d-phenylalanine accumulated quickly to reach a high level in R1M tumors. The specificity of this 123I-2-iodo-d-phenylalanine uptake by tumors was confirmed by the small contribution of blood flow to tumor activity, by the small but significant amount of radioactivity displaced by l-phenylalanine coupled to the obligatory exchange by LAT1, and by the minor uptake in inflamed tissue.
Moreover, low abdominal background activity, fast clearance from blood, and low uptake in the brain were observed (Figs. 3 and 4; Table 2); these findings are favorable for brain tumor detection as well as peripheral tumor detection. Compared with those for 2-iodo-l-phenylalanine, the net tumor uptake and RTB for 2-iodo-d-phenylalanine were similar, but the d-isomer was cleared from blood 2 times faster (Figs. 1 and 3) (3). The latter properties will result in better ratios of uptake in tumors to uptake in blood and less radiation burden to the animal.
Exogenous uptake of d-amino acids (by ingestion or from microorganisms) will lead to metabolism by d-amino acid oxidase, present in the liver, kidneys, and pancreas parenchyma. The latter, together with the stereospecificity of the translational machinery, will result in the formation of proteins uniquely consisting of l-amino acids (10,19). Although 2-iodo-d-phenylalanine showed high uptake in the pancreatic zone, it was excreted very fast. This observation could imply that the d-isomer, just like 123I-2-iodo-l-phenylalanine, is not incorporated into cell proteins, because the pancreas in rodents demonstrates high protein turnover. In humans, high uptake of radiolabeled amino acids in the pancreas does not occur (10).
The above-mentioned characteristics of 123I-2-iodo-d-phenylalanine make this tracer a potential diagnostic tool for oncologic imaging. The rapid clearance from blood, together with the high, fast, and specific tumor uptake, should lead to specific targeting of the tumor with minor radiation burden to other organs and tissues, leading to the possible therapeutic application of this tracer.
Relative to the DPI study, the dissection study presented more detailed, but similar, results regarding tracer distribution. It confirmed the high tumor uptake but the low abdominal uptake of 123I-2-iodo-d-phenylalanine and its renal clearance. Regarding dosimetry, important differences among the 3 amino acid analogs were observed; of the 3 tracers, 2-iodo-l-tyrosine showed the highest effective dose, but 2-iodo-l-phenylalanine and 2-iodo-d-phenylalanine showed comparable doses. Moreover, accumulation in the pancreas is typical in rodents but does not appear in humans, a property that will additionally lead to lower effective doses (e.g., 2.61 × 10−2 for the d-analog). All 3 amino acid analogs showed a favorable biodistribution for a tumor imaging agent. The effective dose will lead to a radiation dose that is comparable to those used in other diagnostic nuclear medicine procedures if the trend seen in the mouse model is seen in humans. Indeed, administration of the compounds to healthy volunteers in a study of human biodistribution will result in an effective dose of less than 1 mSv, the limit for category IIb studies (20).
By use of the hierarchical classification of the organs with the experimental fitted kinetic parameters obtained for 123I-2-iodo-d-phenylalanine, 123I-2-iodo-l-phenylalanine, and 123I-2-iodo-l-tyrosine, a first step toward physiologic modeling could be undertaken. This analysis indicated that not every organ should be considered separately in full pharmacokinetic modeling because of similar pharmacokinetic behaviors within the groups. Five different groups could be defined: tumor, remainder of the body, stomach, blood group, and other-organ group. The stomach as an individual group could be explained by the uptake of free 123I− after dehalogenation.
The above-mentioned characteristics of 123I-2-iodo-d-phenylalanine make this tracer a potential diagnostic tool for oncologic imaging, similar to the l-analog but with the potential for therapeutic application. The rapid clearance from blood, together with the high, fast, and specific tumor uptake, should lead to specific targeting of the tumor with minor radiation burden to other organs and tissues.
CONCLUSION
123I-2-Iodo-d-phenylalanine is a promising tracer for diagnostic oncologic imaging because of its high, fast, and specific tumor uptake and fast clearance from blood. Moreover, the new tracer possesses better diagnostic imaging characteristics regarding enhanced tumor contrast than its l-analog.
Footnotes
Received Jun. 28, 2005; revision accepted Sep. 14, 2005.
For correspondence or reprints contact: Veerle Kersemans, MSc, 804-925 Bay St., Toronto M5S 3L4, Ontario, Canada.
E-mail address: veerle.kersemans{at}utoronto.ca